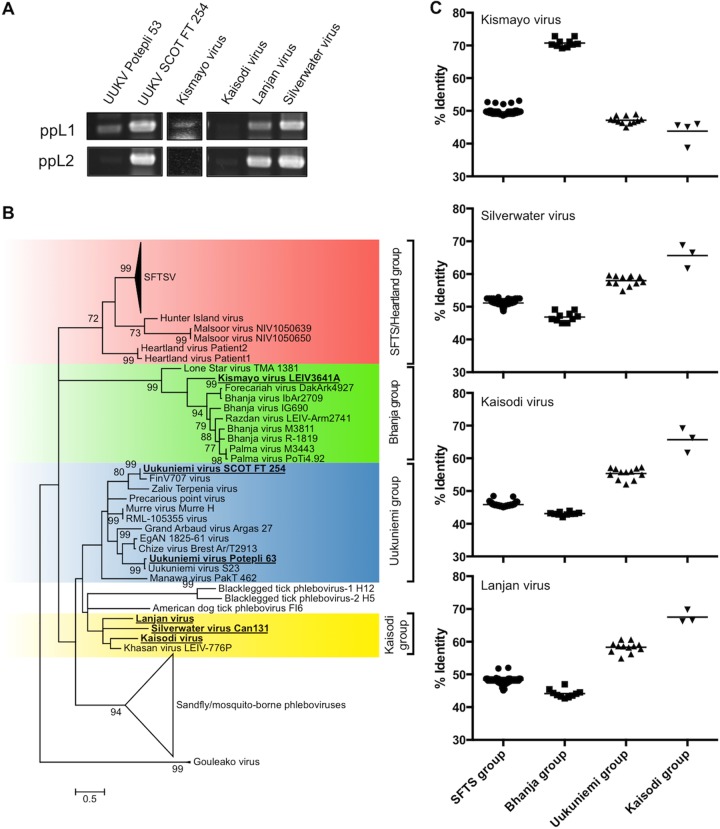
FIG 3.
Retrospective identification of TBPVs. (A) Conventional RT-PCR was performed using RNA from uncharacterized bunyaviruses obtained from multiple sources. (B) Each amplified RT-PCR fragment (around 500 bp) was sequenced, and a phylogenetic tree was constructed based on the fragment sequences and reference sequences using the maximum likelihood (ML) method with 1,000 bootstrap replicates. Bootstrap probabilities above 70% are shown near the branches. The viruses that were newly sequenced in the present study are shown in boldface type and underlined. (C) The sequence identity of the fragment between the newly identified TBPVs (i.e., Kismayo virus, Silverwater virus, Kaisodi virus, and Lanjan virus) and known TBPVs. Each symbol shows the identity value for an individual tick, and the median value for the genetic group is shown as a black bar. Abbreviations: UUKV, Uukuniemi virus; SFTSV, severe fever with thrombocytopenia syndrome virus.