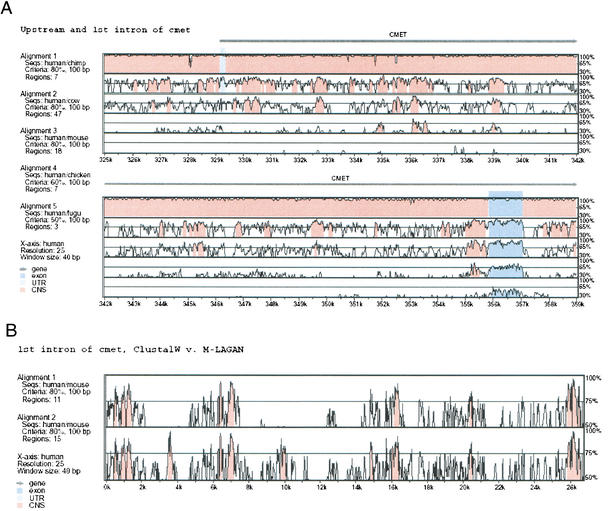
Figure 2.
Visualization of a multiple alignment using VISTA. (A) MLAGAN alignments can be visualized using VISTA, if they are projected to pairwise alignments with respect to one reference sequence. This plot shows the conservation between human and chimpanzee, cow, mouse, and fugu around the first intron of the cMet gene. The human/chimpanzee conservation is uniformly very high; human/cow and human/mouse show varying levels of conservation. The human/chicken alignment also shows some conservation in the non-coding areas. The human/fugu alignment shows conservation only within the first coding exon, and to a lesser degree within the regions upstream and downstream of that exon. (B) First introns of cMet, comparison of CLUSTALW and MLAGAN alignments. We compared the alignment generated by LAGAN and CLUSTALW for the first intron of the cMet gene in eight mammalian sequences (human, baboon, cat, dog, cow, pig, mouse, and rat). The alignments between all of the species except rodents were similar. VISTA plots of the projections to human and mouse are shown. CLUSTALW (top) misaligned the mouse sequence around 4 Kb and 10 Kb, whereas MLAGAN (bottom) found significant conservation in these regions.