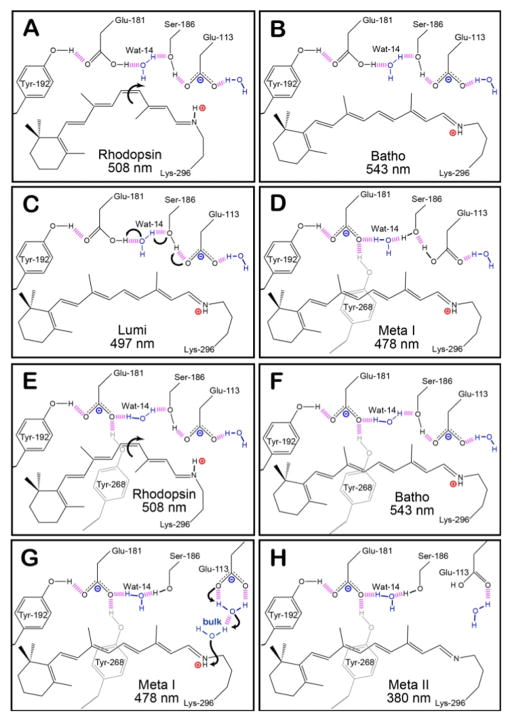
Figure 1.
The hydrogen-bonding network of the protein-binding site of native rhodopsin at select intermediates during the photobleaching sequence. Panels A–D depict the original model of the counter-ion switch, in which Glu-181 is initially neutral. The primary photochemical event involves the isomerization of 11-cis retinal in rhodopsin (A) to all-trans retinal in Batho (B). During these early intermediates, Glu-113 serves as the primary counter-ion for the PSB. The transition from Lumi (C) to Meta I (D) is characterized by the transfer of a proton from Glu-181 to the hydrogen bonding network, which subsequently leads to the protonation of Glu-113. Panels E–H demonstrates our model, [42] which predicts that Glu-181 is also negatively charged during the early intermediates (E and F), and the hydrogen-bonding network rearranges to allow Glu-181 to serve as the primary counter-ion after the transition to Meta I (G). During the Meta II state (H), the PSB is deprotonated within the protein-binding site and the protein is activated in order to catalyze the visual transduction cascade. The purple dashed lines represent hydrogen bonding, and the positively and negatively charged species are indicated using red and blue labels, respectively.