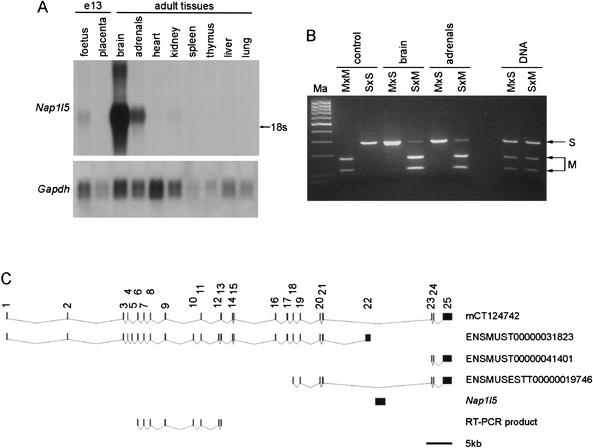
Figure 2.
Tissue-specific and imprinted expression of Nap1l5. (A) Northern blot hybridization of total RNA from adult tissues and day-e13 foetus and placenta, using an antisense RNA probe from within the sequence of AB041556. The size of the transcript on the Northern blot is, in comparison to 18S/28S rRNA, consistent with the expected full-length transcript sequence. Hybridization with Gapdh gives an indication of the amount of RNA loaded. (B) RT-PCR/RFLP analysis of imprinted expression of Nap1l5 in hybrids and backcross animals between Mus musculus C57BL/6 (M) and M. spretus (S). BstZI specifically digests PCR products from the M. musculus allele and assigns the parental origin of alleles in RT-PCRs from reciprocal hybrid RNA. Samples analyzed are brain and adrenal gland RNAs from a cross between C57BL/6 female and M. spretus male (M × S, maternal inheritance of M allele, paternal inheritance of S allele) and from a cross between an F1 (C57BL/6 × M. spretus) female and C57BL/6 male (S × M, maternal inheritance of S allele, paternal inheritance of M allele at this locus). The DNA amplification indicates the presence of both alleles in the animals examined. No product amplified in control reactions with no reverse transcriptase (not shown). The controls are amplified M. musculus C57BL/6 (M × M) or M. spretus (S × S) DNA to indicate the digestion products. Ma is a 100-bp molecular weight marker. (C) Map indicating the position of Nap1l5 within a large intron of Herc3. mCT124742 is a transcript described in the Celera database; ENSMUST00000031823, ENSMUST00000041401, and ENSMUSEST00000019746 are Ensembl identifiers. mCT124742 and ENSMUSEST00000019746 indicate that an intron of Herc3 transcript spans the region containing Nap1l5; Herc3 and Nap1l5 are transcribed in the opposite orientation. The RT-PCR product used to investigate imprinting of Herc3 in T77H uniparental duplication tissues is shown.