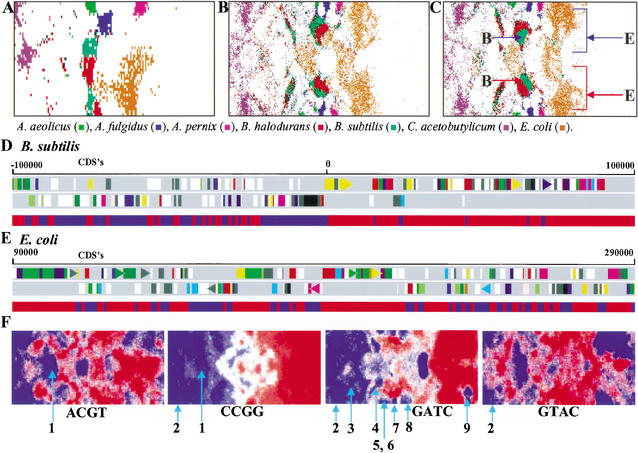
Figure 3.
Intraspecies separations and tetranucleotide distributions in SOMs for bacterial genomes. (A,B,C) The 10-kb tri-, 1-kb tri-, and 1-kb tetra-SOMs. Seven representative species with two major zones are indicated in color as detailed in the figure above. In C, the two major zones of B. subtilis or E. coli are noted with red or blue arrows with the letter B or E, respectively. (D) Transcriptional polarity and SOM separation for B. subtilis sequences. Two transcriptional polarities of CDSs in the 200-kb B. subtilis segment with a replication origin are presented separately in the top two panels; this was obtained from the DDBJ Web site (http://gib.genes.nig.ac.jp/). Below the two panels, contiguous 1-kb sequences within the 200-kb segment and belonging to the two major zones marked with red and blue arrows in C are shown separately with the red and blue bands, respectively. (E) Transcriptional polarity and SOM separation for E. coli sequences. A 200-kb E. coli segment lacking a replication origin was analyzed as described in D. (F) Tetranucleotide distribution in the 1-kb tetra-SOM for bacteria. Levels of each tetranucleotide for all lattice vectors in the tetra-SOM of Fig. 2C were divided into five categories containing an equal number of lattices, and the highest, second-highest, middle, second-lowest, and lowest categories are shown with different levels of red and blue as described in Fig. 1,D–F. Zones for bacteria that have genes encoding a restriction enzyme that recognizes the respective tetranucleotide are noted by light blue lines with the following numbers to show the species name: (1) H. pylori; (2) M. jannaschii; (3) S. aureus; (4) S. pneumoniae; (5) P. abyssi; (6) P. horikoshii; (7) A. fulgidus; (8) A. pernix; and (9) D. radiodurans. For other palindromic tetranucleotides, see Supplementary Data 2. Of 17 restriction enzymes from 11 bacteria, the respective tetranucleotides were under-represented in 15 instances.