Figure 2.
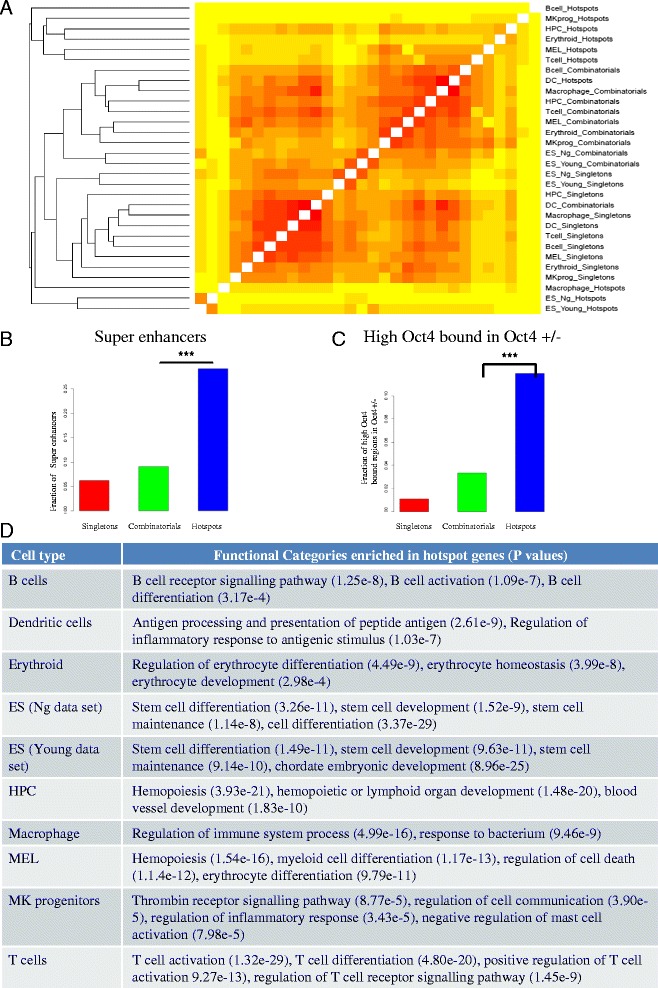
Hotspots preferentially bind in cell type-specific gene neighbourhoods. A. A heatmap of Pearson’s correlation coefficients of genes in 30 sets (three groups of 10 cell types) with a hierarchical clustering tree showing that singletons and combinatorials clustered together in a tight cluster while hotspot gene sets were very distinct from each other. B. Fraction of ES super enhancers defined by Whyte et al. [11] in three groups showing hotspots significantly overlapped with super enhancers. C. Fraction of high Oct4 bound regions in Oct4+/− compared to Oct4+/+ [12] ES cells in three groups showing hotspots enriched for high Oct4 bound regions in Oct4+/− and also pluripotent genes D. Functional enrichments of hotspot genes in 10 cell types along with p values showing that hotspot genes were enriched for cell type specific genes.
