Figure 5.
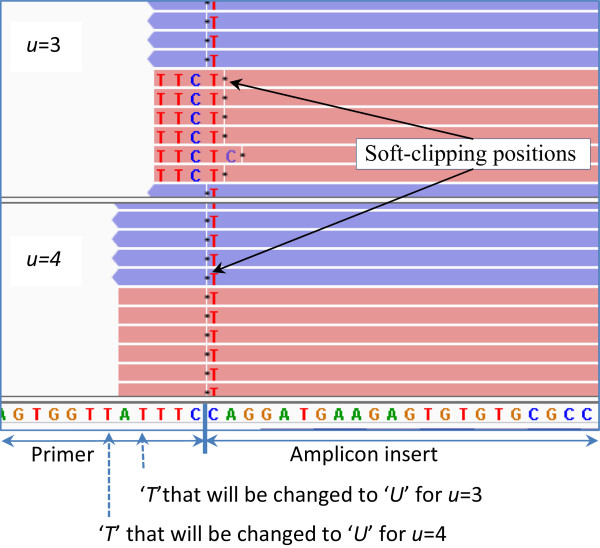
An instance of misalignment at u = 3 which is corrected for larger values of u . The top panel shows simulated reads with u = 3, and the bottom panel shows simulated reads with u = 4. The reads in the forward direction are in pink and the reads in the reverse direction are in blue. A C- > T mutation (highlighted) has been inserted into some of the reads at the first position in the insert. When u = 3, only three bases of the primer are left in the read after primer digestion. The C- > T mismatch causes the alignments of all the forward reads to be clipped to exclude the variant from the alignment. Breaks in the reads indicate soft-clipping positions. At u = 4, five bases of the primer are left, which was sufficient to avoid the misalignments.
