Figure 3.
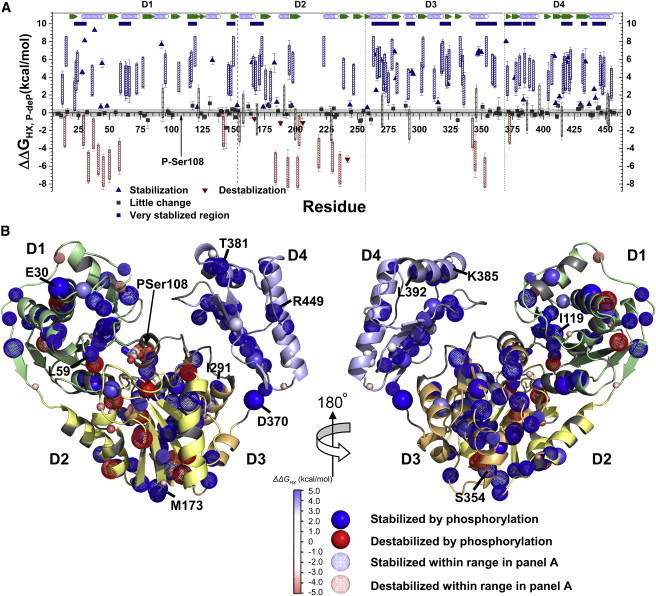
Phosphorylation stabilizes or slows HX at more sites than it accelerates. (A) Rate constants of HX, kex, measured either as HDX by NMR or on the subsecond scale by CLEANEX-PM (29), were transformed to ΔGHX using Eq. 2 and the ΔΔGHX,P-deP differences are plotted. Blue indicates where phosphorylation is significantly stabilizing, and red indicates where it is destabilizing. Triangles represent cases in which kex was measured in both Apo-P and Apo-deP forms. Hatched bars mark cases in which kex in one phosphorylation state lay in the intermediate, unmeasured time range where 1 s−1 > kex > 4 × 10−3 s−1; the height of the bar indicates ΔΔGHX,P-deP lies within this range. Sequence segments with more than four of 10 residues protected by phosphorylation >2 SD above average are marked with navy blue bars. Locations of strands and helices are given in green and open blue symbols, respectively. (B) Amide groups protected from HX by phosphorylation of Ser-108 are marked by spheres with shades of blue, and amide groups destabilized by phosphorylation are in red. Dotted spheres correspond to residues with the uncertainty ranges marked in (A). The amplitude of the phosphorylation-dependent change, ΔΔGHX, is symbolized by the color intensity and radii of the spheres. To see this figure in color, go online.
