Figure 1.
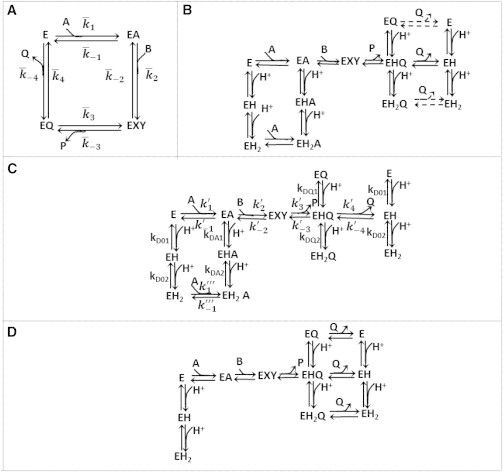
Schematic of the models employed. (A) An ordered bi-bi mechanism without any pH dependency; denotes the apparent rate constants. (B) Ordered bi-bi mechanism with proposed pH mechanism; it is assumed that the coenzymes bind to all charged states. (Dashed lines) Reaction steps for which the rate constants are not identifiable for cMDH-catalyzed reaction. (C) Proposed model for cMDH. NAD binds to unprotonated and di-protonated enzyme states, while NADH binds to the uniprotonated enzyme state. (D) Proposed mechanism for pH dependency for mMDH (15). (Arrows) Binding/dissociation of coenzyme/substrate. In each of these schematics, A, B, P, and Q represent NAD, MAL, OAA, and NADH, respectively. In these schematics, EXY denotes the rapid conversion of EAB to EPQ.
