Figure 4. FL suppresses REs associated with GC identity.
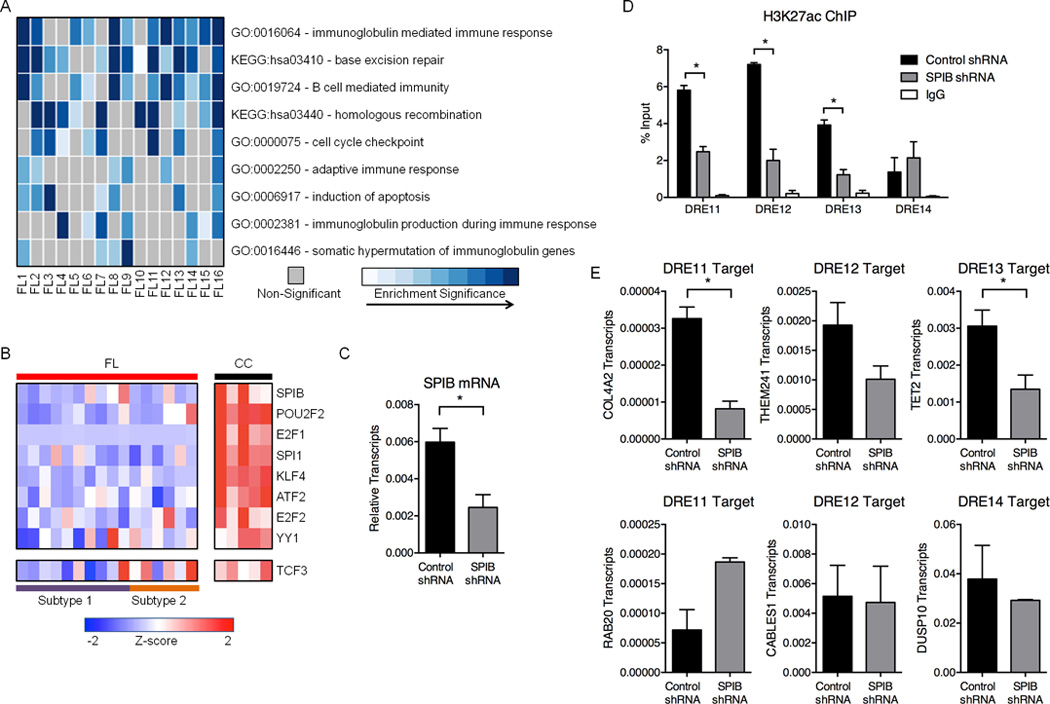
A) Heatmap representation of enriched Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways identified from down-regulated genes within 500kb of attenuated DREs. B) Expression profiles (microarray) for TFs predicted to bind motifs in attenuated DREs. Refer to Table S6 for expression values. C) SPIB transcripts, as measured by qPCR, in GM12878 cells transfected with either control or SPIB shRNA. Results represent the mean ± SEM of three independent experiments. D) H3K27ac ChIP assays in GM12878 cells transfected with control or SPIB-specific shRNA. Associated DNA was analyzed via qPCR using primers spanning DREs harboring SPIB binding sites. Results represent the mean ± SEM of three independent experiments. E) Transcript levels for DRE target genes, as measured by qPCR, in GM12878 cells transfected with either control or SPIB-specific shRNA. Results represent the mean ± SEM of three independent experiments. Statistical significance (unpaired t-test): * p≤ 0.05.
