Figure 7. Polymorphisms and mutations in FL-altered DREs disrupt enhancer function.
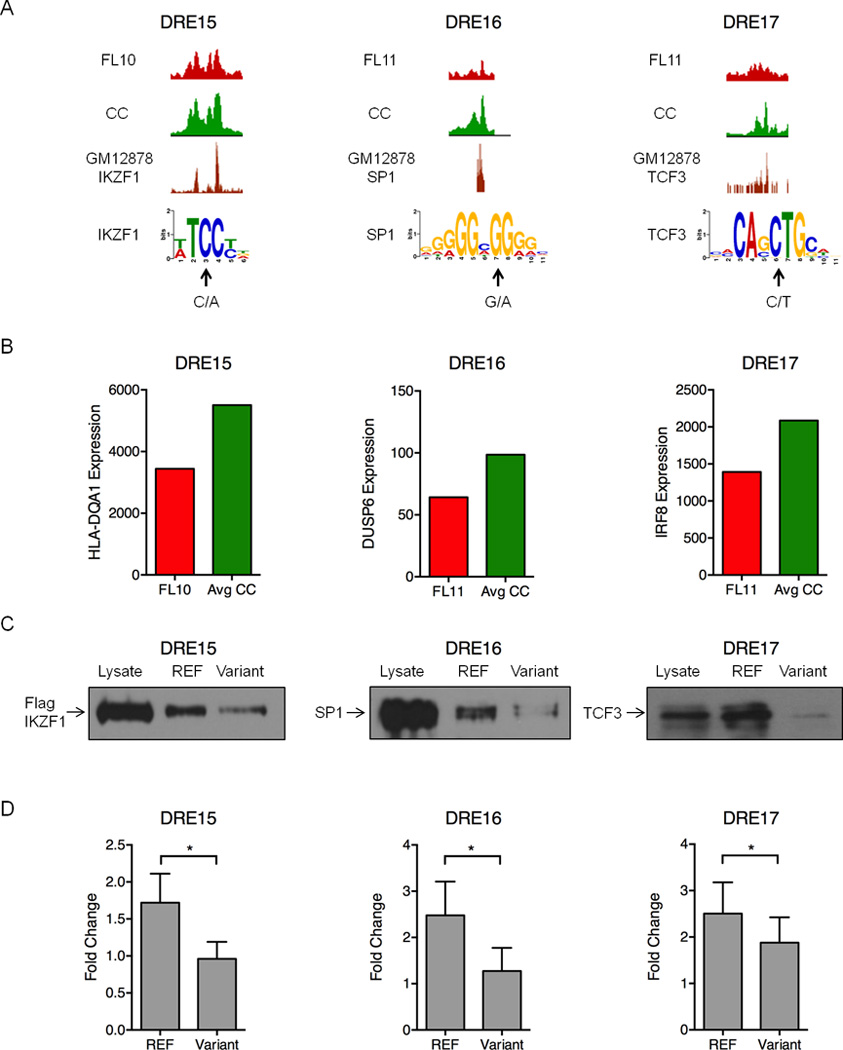
A) UCSC Genome Browser views of H3K27ac ChIP-seq data illustrating attenuated DREs harboring the indicated sequence variants in FL and the reference sequence in CC (upper 2 tracks). The bottom track shows ChIP-seq data for the indicated TFs performed in the GM12878 B cell line (Neph et al., 2012). Arrows indicate the variant sequence and location in PWMs for each TF. B) Expression of the DRE target genes quantified by microarray analysis. C) Oligonucleotide precipitation assays demonstrate reduced TF binding in variant-containing compared to reference sequences. Western blots were probed with antibodies specific to the TF of interest (representative of three experimental replicates). D) Luciferase reporter assays performed in lymphoma cell lines demonstrate significantly reduced activity for the variant-containing compared to reference sequences in the attenuated DREs. Luciferase activity is presented as a fold change for the enhancer vector relative to a reporter containing only the SV40 promoter (set to a value of 1.0). Results represent the mean ± SEM of three independent experiments. Statistical significance (paired t-test): * p≤ 0.05.
