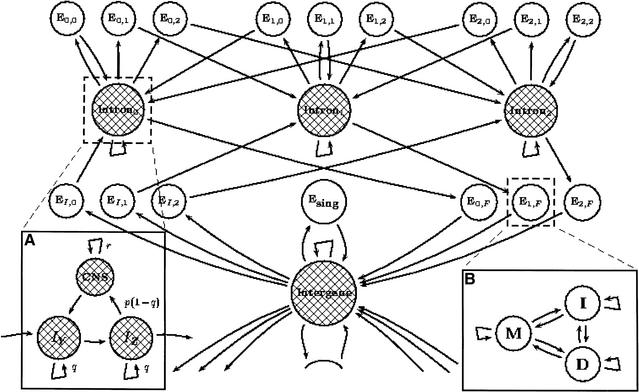
Figure 2.
A GPHMM for alignment and prediction of exons using genomic DNA from two different organisms. The shaded states are the typically less-conserved intergene and intron states, each producing either a single base or a gap in each organism. The use of self-transitions models their state durations as geometric. The unshaded states (all of which are exons) will all have duration 1, as they have no self-transitions; however, they are generalized and produce exon pairs according to some predetermined joint distribution. (A) In order to avoid the prediction of coding exons in all conserved regions, it was necessary to introduce conserved noncoding states (CNS). Each intron and intergene state consist of two parts: an I-state for modeling long unrelated noncoding regions, and a CNS state for modeling interspersed conserved domains. (B) The modeling of coding exon states in pairs required the construction of a specialized PHMM, consisting of match/mismatch (M), insertion (I), and deletion states (D), which was used to assign probabilities to exon pairs based on alignments in protein space using an appropriate evolutionary model.