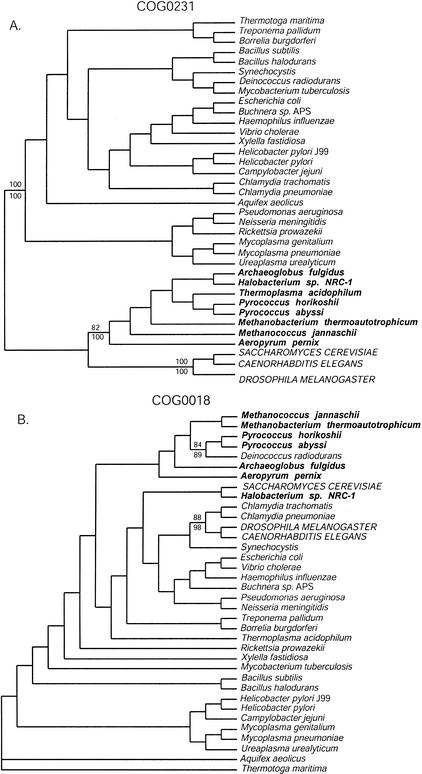
Figure 1.
Examples of three-domain and non-three-domain phylogenetic trees from analyses of the COG database protein alignments. The trees are the single shortest trees found by a maximum parsimony (MP) analysis of the amino acid alignments (neighbor-joining [NJ] analysis gave the same topology). Names of organisms belonging to the Bacteria are in italics; names of Archaea are in bold italics, and names of Eucarya are in all capital letters. (A) The phylogeny of COG0231 (efp in E. coli) recapitulates the basic three-domain topology given by ribosomal RNA, and the numbers indicate bootstrap support for the monophyly of the archaeal, bacterial, and eucaryal sequences in this COG. Results from MP bootstrap analysis are given above the branches, and results from NJ bootstrap analysis are given below. (B) The phylogeny of COG0018 (argS in E. coli) violates the three-domain paradigm, as none of the three domains are monophyletic. Indications of horizontal gene transfer events are presented in enlarged font along with corresponding bootstrap support for the nodes demarking lateral transfer.