Abstract
Alcohol binge-drinking during adolescence is a serious public health concern with long-term consequences. We used RNA sequencing to assess the effects of excessive adolescent ethanol binge-drinking on gene expression in the dorsal raphe nucleus (DRN) of alcohol preferring (P) rats. Repeated binges across adolescence (three 1h sessions across the dark-cycle per day, 5 days per week for 3 weeks starting at 28 days of age; ethanol intakes of 2.5 – 3 g/kg/session) significantly altered the expression of approximately one-third of the detected genes. Multiple neurotransmitter systems were altered, with the largest changes in the serotonin system (21 of 23 serotonin-related genes showed decreased expression) and GABA-A receptors (8 decreased and 2 increased). Multiple neuropeptide systems were also altered, with changes in the neuropeptide Y and corticotropin-releasing hormone systems similar to those associated with increased drinking and decreased resistance to stress. There was increased expression of 21 of 32 genes for potassium channels. Expression of downstream targets of CREB signaling was increased. There were also changes in expression of genes involved in inflammatory processes, axonal guidance, growth factors, transcription factors, and several intracellular signaling pathways. These widespread changes indicate that excessive binge drinking during adolescence alters the functioning of the DRN and likely its modulation of many regions of the central nervous system, including the mesocorticolimbic system.
Keywords: serotonin, GABA-A receptors, neuropeptides, CREB, ion channels, inflammation, RNA sequencing, alcoholism
1. Introduction/Background
A multi-national review of studies of adolescent drinking indicates that about ¾ of adolescents have consumed alcohol by late adolescence, and 20–40% had engaged in binge drinking, the consumption of 5 or more drinks on one occasion (Marshall, 2014). In the US, adolescents (ages 12–20) drink 11 percent of all alcohol consumed, 90% of which is consumed during binge drinking (NIAAA, 2013).
Adolescence is a time in which brain connections mature and are remodeled, and alcohol could alter these processes. Given the legal and ethical prohibition of using adolescents in alcohol research, animal models have been used to examine the effects of ethanol on adolescents. Studies have examined the effects of chronic low dose exposure (Evrard et al., 2006) and different models of binge drinking (McBride et al., 2014; Pascual et al., 2007; Pascual et al., 2014; Ward et al., 2014). Selectively bred alcohol preferring (P) and high-alcohol-drinking (HAD) rats engage in binge-like drinking, and adults and peri-adolescents of both sexes readily achieve blood ethanol levels ≥ 80 mg% (Bell et al., 2014). Multiple scheduled access protocols (usually three 1 h periods of access to alcohol during the dark phase) enhance this binge-like drinking, and under these conditions peri-adolescent selectively bred rats consume more alcohol than their adult counterparts (Bell et al., 2014). In general, animal studies and human observational studies during adolescence indicate that adolescents, relative to adults, are less affected by ethanol-induced sedation and motor incoordination but more affected by its rewarding and reinforcing properties (Bell et al., 2013; Spear, 2010). Greater disturbances in some cognitive functions have also been reported (Bell et al., 2013; Spear, 2010).
Alcohol dependence has two basic components: positive effects primarily attributed to ethanol stimulated release of dopamine in the reward centers of the brain and negative effects of anxiety and depression that occur after cessation of drinking (Koob, 2013). The reward system in the brain includes the mesocorticolimbic dopamine system and extended amygdala, which intersect in the nucleus accumbens shell. Within this system, ethanol increases the release of dopamine in the shell of the nucleus accumbens, which is directly associated with the rewarding effects of ethanol consumption (McBride, 2002; McBride and Li, 1998) reviewed in Koob (2013). Serotonin modulates the effect of ethanol on dopamine release via the 5-HT3 receptor (Campbell et al., 1996; Campbell and McBride, 1995; Engleman et al., 2008; Wozniak et al., 1990).
In rats, a genetic predisposition for excessive ethanol consumption is associated with alterations in central serotonergic activity. For example, ethanol-naïve P and HAD rats have significantly lower serotonin and/or 5HIAA5-hydroxyindoleacetic acid levels in the frontal cortex, nucleus accumbens, caudate putamen, hippocampus and hypothalamus compared with ethanol-naïve NP and LAD rats (Bell et al., 2012; Gongwer et al., 1989; Murphy et al., 1987). In addition, ethanol-naïve P, and in some cases ethanol-naïve HAD, rats have altered levels of 5HT1A, 5HT1B, 5HT2, 5HT2C and 5HT3 receptors in multiple brain regions compared with ethanol-naïve NP and LAD rats (McBride et al., 1997a; McBride et al., 1997b). Thus, alterations in receptor expression may be compensating for reduced serotonin function. Selective agonists and/or antagonists for various serotonin receptors affect the alcohol-consuming behavior of rats selected for high alcohol preference (Ding et al., 2012; Lankford et al., 1996; Lankford and Myers, 1996; Long et al., 1996; Overstreet et al., 1997; Rodd-Henricks et al., 2000; Rodd et al., 2010). Other selectively bred high alcohol-consuming rat lines (Alko Alcohol preferring and Sardinian alcohol preferring rats) had higher levels of serotonin in whole brain or selected regions than their low alcohol-consuming counterparts (Alko Non-Alcohol and Sardinian non-preferring rats) (Bell, Sable et al. 2012).
The dorsal raphe nucleus (DRN) is an origin of the central serotonergic system. About 50% of the serotonin neurons in the rat brain are in the DRN, although fewer than 50% of the neurons in the DRN are serotonergic (Waselus et al., 2011). The DRN has projections into numerous brain regions, including the mesolimbic regions important in reward and addiction (amygdala, hippocampus, caudate putamen, substantia nigra and ventral tegmental area). Projections from the raphe nuclei mediate dopamine release in the ventral tegmental area (Liu et al., 2006b; Rodd et al., 2007; Sari et al., 2011). Ethanol-naïve P rats have fewer serotonergic neurons in the DRN than ethanol-naïve NP rats and decreased serotonergic projections into terminal regions (Zhou et al., 1994). Postmortem studies of the DRN of alcoholics (Underwood et al., 2007) found less serotonin transporter in the brainstem of alcoholics.
The links between the DRN and the reward centers of the brain, its role in anxiety, stress and depression, and the changes that occur during adolescence make it an important region in which to study the effects of heavy alcohol exposure, particularly during the vulnerable period of adolescence. This study examines the effects of binge drinking by adolescent P rats on the gene expression profile of the DRN.
2. Materials and Methods
2.1 Animal Binge drinking protocol
Peri-adolescent male P (alcohol preferring) rats were maintained on a reverse light-dark cycle and given free-choice access to ethanol using a multiple-scheduled-access procedure, as described in Bell et al. (2011). There were 10 animals in the water control group and 11 in the ethanol binge-drinking group. Animals were given ad libitum access to food and water and those in the drinking group were given access to ethanol (15 and 30% ethanol solutions concurrently) in 3 x 1-h sessions per day for 5 consecutive days/week, starting at 28 days of age. The three 1hr ethanol access sessions were scheduled so that the first 1hr session started at dark onset (1000 h), the second 1 h session began 2 h after the end of the first session (1300 h) and the third/last 1hr session began 2 h after the end of the second session (1600 h). The animals used in this study are the same ones described in McBride et al. (2014). A graph of the 15-day ethanol drinking data is given in McBride et al. (2014). In general, during week 1, ethanol intakes ranged from 3–4 g/kg/1hr session with a total intake of approximately 10 g/kg/day; during weeks 2 and 3, ethanol intakes ranged from 2–3 g/kg/1hr session with a total intake of approximately 8 g/kg/day.
The rats were sacrificed 3 h after the 1st access session on their 15th day of drinking. Brains were rapidly extracted and flash-frozen in isopentane in dry ice and stored at −80 C until sectioning. Brains were sectioned (300 μm) and the DRN micro-punched using procedures previously described (McBride et al., 2014). The DRN was punched from coronal sections between −7.30 mm to −8.30 mm post bregma according to the rat brain atlas by Paxinos and Watson (1998). All equipment used to obtain tissue was treated with RNase Zap (Life Technologies, Carlsbad, CA). Other brain regions of these animals have been studied (McBride et al., 2014). The animals used in these experiments were maintained in facilities fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). All research protocols were approved by the institutional animal care and use committee and are in accordance with the guidelines of the Institutional Care and Use Committee of the National Institute on Drug Abuse, National Institutes of Health, and the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council 1996).
2.2 Sample collection and quality
To extract the RNA, punches were immediately homogenized in Trizol (Life Technologies, Carlsbad, CA) and processed according to manufacturer’s protocol but with twice the suggested ratio of Trizol to tissue (Edenberg et al., 2005). Ethanol-precipitated total RNA was further purified through Qiagen RNeasy columns (Qiagen, Hilden Germany) according to the manufacturer’s protocol. The yield, concentration and purity of the RNA were measured by Nanodrop (Thermo Fisher Scientific, Waltham, MA) spectrum from 220 nm to 340 nm. Quality was also assessed by Agilent Bioanalyzer (Agilent Technologies, Santa Clara, Ca); RNA integrity numbers (RIN) averaged 8.6 for the samples, showing little degradation.
2.3 RNA sequencing
Standard methods were used for RNA-seq library construction, EZBead preparation and Next-Gen sequencing using the Life Technologies SOLiD system. Briefly, total RNA (≤ 400 ng per sample) was first ribosome-depleted using Ribominus™ Eukaryote Kit for RNA-Seq (Ambion, CA), and whole transcriptome library was prepared and barcoded per sample using the standard protocol of SOLiD Total RNA-seq Kit (Life Technologies, Carlsbad, CA). Each barcoded library was quantified by quantitative PCR using SOLiD Library Taqman qPCR Module (Life Technologies, Carlsbad, CA), and pooled in equal molarity. EZBead preparation, bead library amplification, and bead enrichment were then conducted using Life Technologies EZ Bead™ E80 System (Life Technologies, Carlsbad, CA). And finally sequencing by ligation was carried out using standard single-read, 5′-3′ strand-specific sequencing procedure on SOLiD4™ Sequencer (50b-read), as well as on SOLiD™ 5500xl Sequencer (75 b read). The average number of mapped reads per sample was 20.8 million.
2.4 Data Processing and Quality Assessment
We used SOLiD Instrument Control Software and SOLiD Experiment Tracking System software for the read quality recalibration. Each sequence read was scanned for low-quality regions, and if a 5-base sliding window had an average quality score less than 20, the read was truncated at that position. Any read with a length of less than 35 bases was discarded. Our experience suggests that this strategy effectively eliminates low-quality reads while retaining high-quality regions (Breese and Liu, 2013; Juan et al., 2013; Todd et al., 2013).
2.5 Sequence Alignment
We used BFAST (http://bfast.sourceforge.net) (Homer et al., 2009) as our primary alignment algorithm because it has high sensitivity for aligning the reads on the loci containing small insertions and deletions compared to the reference genome (rn4). We used a TopHat-like strategy (Trapnell et al., 2009) to align the sequencing reads that cross splicing junctions using NGSUtils (http://ngsutils.org/) (Breese and Liu, 2013). After aligning the sequence reads to a filtering index including repeats, ribosome RNA, and other sequences that are not of interest, we conducted a sequence alignment for three levels: genome, known junctions (University of California Santa Cruz Genome Browser), and novel junctions (based on the enriched regions identified in the genomic alignment). We restricted our analysis to the uniquely aligned sequences with no more than two mismatches.
2.6 Analysis
Differential Expression between the drinking and water groups was determined using the R package EdgeR with continuous dispersion (Robinson et al., 2010). FDR was calculated according to Benjamini and Hochberg (1995).
Differentially expressed genes were analyzed using the Winter 2013 Release of QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). Pathway, Functional and Upstream analysis were performed.
2.7 Convergence Analysis
We assembled a list of genes that were identified in GWAS studies (Bierut et al., 2010; Edenberg et al., 2010; Hack et al., 2011; Johnson et al., 2011; Kendler et al., 2011; Lind et al., 2010; Treutlein et al., 2009; Wang et al., 2013; Zlojutro et al., 2011; Zuo et al., 2012) and postmortem studies of several brain regions. The human post-mortem studies examined superior frontal cortex (Lewohl et al., 2000; Liu et al., 2007; Liu et al., 2006), frontal cortex (Liu et al., 2007; Mayfield et al., 2002), prefrontal cortex (Flatscher-Bader et al., 2005; Iwamoto et al., 2004), temporal cortex (Sokolov et al., 2003), motor cortex (Mayfield et al., 2002) nucleus accumbens and ventral tegmental area (Flatscher-Bader et al., 2010; Flatscher-Bader et al., 2005), basolateral amygdala (Kryger and Wilce, 2010) and hippocampus (McClintick et al., 2013; Zhou et al., 2011).
The animal gene expression studies used for comparison were all P/NP studies done by our group. These included comparisons between naïve P and NP animals, as well as P animals exposed (or not) to alcohol using various models of drinking, different ages and gender. These previous studies predominantly used the hippocampus, ventral tegmental area, accumbens, amygdala or sub-regions of these areas. (Bell et al., 2009; Kimpel et al., 2007; McBride et al., 2013a; McBride et al., 2012; McBride et al., 2013b; McBride et al., 2010; Rodd et al., 2008). None examined the DRN. Genes from microarray studies using the central amygdala and the accumbens shell from these same animals (McBride et al., 2014) are noted specifically. Genes from these 3 types of analyses (GWAS, post-mortem human and P/NP animal studies) were matched to the DRN data by gene symbol.
3. Results
3.1 RNA-seq results
We used RNA-seq to examine the pattern of gene expression in the DRN of peri-adolescent male P rats that had consumed large amounts of ethanol in a repeated binge pattern over a 3-week period (post-natal days 28 to day 49). Post-natal day 28 coincides with the beginning of adolescence, which ends at approximately day 42 in female rats and day 45 in male rats (Bell et al., 2013; Spear, 2010). The average consumption was approximately 8 g/kg per day for the 5 drinking days of each week (McBride et al., 2014). These levels of intake meet the criterion for binge-drinking put forth by the National Institute on Alcohol Abuse and Alcoholism (NIAAA, 2004), blood ethanol levels ≥ 80mg%. There were 12,047 genes detectably expressed in the DRN, of which 3,567 genes (30%) were differentially expressed between ethanol exposed and control animals (at FDR ≤ 0.05; Supplementary Table 1 lists all genes with differential expression). Among the differentially expressed genes, 1391 (39%) had absolute fold changes >1.5 (Figure 1).
Figure 1.
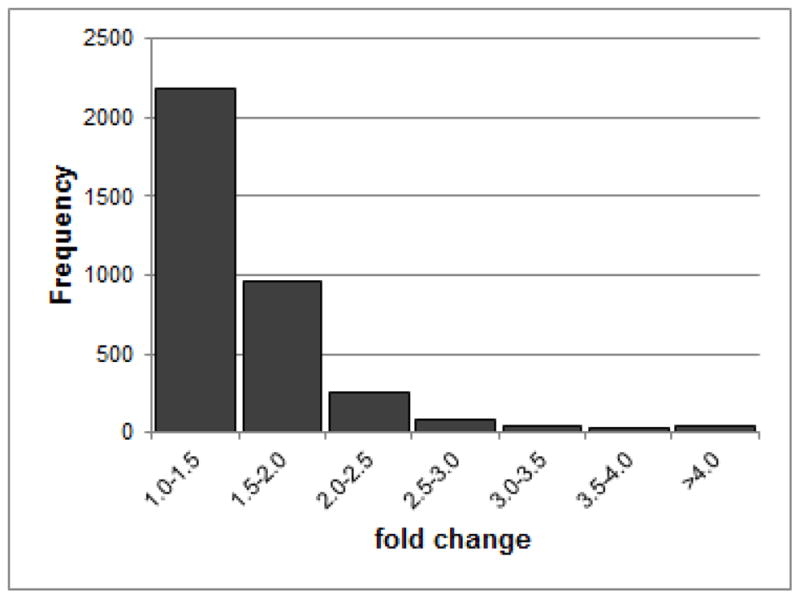
Distribution of fold changes of genes with an FDR ≤ 0.05.
The expression levels of genes encoding biosynthesis and transport of many neurotransmitters were altered, along with genes encoding their receptors (Table 1; Supplementary Table 1). Genes for the serotonin pathway were changed the most (Table 1), with 21 of 23 expressed genes showing decreased expression, including tryptophan hydroxylase, dopa decarboxylase, serotonin transporter Slc6a4, vesicular monoamine transporter Slc18a2 and organic ion transporter Slc22a3. Expression of genes for other neurotransmitter systems was also altered (Table 1). Among the 12 GABAA receptors expressed in the DRN, 8 were decreased and 2 were increased. GABA transporters Slc6a1 and Slc6a13 were also increased. The glutamate system showed decreases in many receptor genes, but large increases in the expression of Grin2c (2.6-fold), Grm1 (2.0-fold) and Grm4 (3.0-fold) as well as increases in glutamate transporters Slc1a6 and Slc1a3. Drd1a was decreased, as were the genes for synthesizing dopamine, the vesicular monoamine transporter VMAT2 (Slc18a2) and several protein phosphatases downstream of dopamine receptors.
Table 1. Neurotransmitter genes differentially expressed.
Neurotransmitter related genes whose expression is altered by adolescent ethanol binge-drinking (FDR<0.05)
| GENE | Fold Change | Gene Name |
|---|---|---|
| Serotonin | ||
|
| ||
| Htr1a | −1.6 | 5-hydroxytryptamine (serotonin) receptor 1A, G protein-coupled |
| Htr1b | −1.28 | 5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
| Htr1d | −1.65 | 5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| Htr2a | −1.4 | 5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| Htr2c | −1.7 | 5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled |
| Htr3a | −1.58 | 5-hydroxytryptamine (serotonin) receptor 3A, ionotropic |
| Htr4 | −1.8 | 5-hydroxytryptamine (serotonin) receptor 4, G protein-coupled |
| Htr5a | −1.48 | 5-hydroxytryptamine (serotonin) receptor 5A, G protein-coupled |
| Htr5b | −2.15 | 5-hydroxytryptamine (serotonin) receptor 5B, G protein-coupled |
| Slc6a4 | −2.06 | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4, also 5-HTT and SERT1 |
| Slc18a2 | −2.1 | solute carrier family 18 (vesicular monoamine), member 2, also VMAT2 |
| Slc22a3 | −2.26 | solute carrier family 22 (extraneuronal monoamine transporter), member 3 (post-synaptic) |
| Maoa | −1.7 | monoamine oxidase A |
| Maob | −1.48 | monoamine oxidase B |
| Tph2 | −2.59 | tryptophan hydroxylase 2 |
| Tph1 | −2.33 | tryptophan hydroxylase 1 |
| Ddc | −2.06 | dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| Gch1 | −2.17 | GTP cyclohydrolase 1 |
| Pcbd1 | −1.68 | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| Pts | -−1.51 | 6-pyruvoyltetrahydropterin synthase |
| Qdpr | −1.56 | quinoid dihydropteridine reductase |
|
| ||
| GABA | ||
|
| ||
| Gabarapl1 | −1.33 | GABA(A) receptor-associated protein like 1 |
| Gabra2 | −1.62 | gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| Gabra3 | −1.69 | gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| Gabra5 | −1.8 | gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| Gabra6 | 4.53 | gamma-aminobutyric acid (GABA) A receptor, alpha 6 |
| Gabrb1 | −1.32 | gamma-aminobutyric acid (GABA) A receptor, beta 1 |
| Gabrd | 4.34 | gamma-aminobutyric acid (GABA) A receptor, delta |
| Gabre | −2.27 | gamma-aminobutyric acid (GABA) A receptor, epsilon |
| Gabrg1 | −1.22 | gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| Gabrg3 | −1.42 | gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
| Gabrq | −2.45 | gamma-aminobutyric acid (GABA) A receptor, theta |
| Gad1 | 1.33 | glutamate decarboxylase 1 (brain, 67kDa) |
| Slc6a1 | 1.27 | solute carrier family 6 (neurotransmitter transporter, GABA |
| Slc6a13 | 1.8 | solute carrier family 6 (neurotransmitter transporter) member 13, also GAT2 |
|
| ||
| Glutamate | ||
|
| ||
| Gria3 | 1.29 | glutamate receptor, ionotropic, AMPA 3 |
| Gria4 | 1.31 | glutamate receptor, ionotropic, AMPA 4 |
| Grid2 | 2.31 | glutamate receptor, ionotropic, delta 2 |
| Grid2ip | 3.49 | glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein |
| Grik3 | 1.35 | glutamate receptor, ionotropic, kainate 3 |
| Grin2b | −1.56 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B |
| Grin2c | 2.61 | glutamate receptor, ionotropic, N-methyl D-aspartate 2C |
| Grin2d | −1.4 | glutamate receptor, ionotropic, N-methyl D-aspartate 2D |
| Grin3a | −1.39 | glutamate receptor, ionotropic, N-methyl-D-aspartate 3A |
| Grinl1a | −1.32 | GRINL1A downstream protein Gdown1 (Grinl1a) |
| Grip1 | −1.36 | glutamate receptor interacting protein 1 |
| Grip2 | −1.58 | glutamate receptor interacting protein 2 |
| Grm1 | 2.06 | glutamate receptor, metabotropic 1 |
| Grm4 | 2.98 | glutamate receptor, metabotropic 4 |
| Grm6 | −1.7 | glutamate receptor, metabotropic 6 |
| Slc1a3 | 1.7 | solute carrier family 1 (glial high affinity glutamate transporter), member 3, also EAAT1 |
| Slc1a6 | 2.79 | solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6, also EAAT4 |
| Slc25a18 | 1.45 | solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| Slc25a22 | 1.3 | solute carrier family 25 (glutamate carrier), member 18 |
|
| ||
| Glycine | ||
|
| ||
| Glra1 | −1.75 | glycine receptor, alpha 1 |
| Glra2 | −2.02 | glycine receptor, alpha 2 |
| Glra3 | −1.56 | glycine receptor, alpha 3 |
| Glrb | −1.24 | glycine receptor, beta |
|
| ||
| Cholinergic | ||
|
| ||
| Chrna4 | −1.48 | cholinergic receptor, nicotinic, alpha 4 (neuronal) |
| Chrna7 | −1.37 | cholinergic receptor, nicotinic, alpha 7 (neuronal) |
| Chrnb3 | −2.44 | cholinergic receptor, nicotinic, beta 3 (neuronal) |
| Slc18a3 | 1.61 | solute carrier family 18 (vesicular acetylcholine), member 3, also VAChT |
|
| ||
| Adrenergic | ||
|
| ||
| Adra1b | −1.59 | adrenoceptor alpha 1B |
| Adra1d | 3.8 | adrenoceptor alpha 1D |
| Adrb2 | 1.4 | adrenoceptor beta 2, surface |
| Adrbk2 | 1.57 | adrenergic, beta, receptor kinase 2 |
|
| ||
| Purinergic | ||
|
| ||
| P2rx2 | −1.85 | purinergic receptor P2X, ligand-gated ion channel, 2 |
| P2rx4 | 1.56 | purinergic receptor P2X, ligand-gated ion channel, 4 |
| P2rx6 | 1.86 | purinergic receptor P2X, ligand-gated ion channel, 6 |
| P2ry12 | −1.36 | purinergic receptor P2Y, G-protein coupled, 12 |
|
| ||
| Dopamine | ||
|
| ||
| Drd1a | −1.36 | dopamine receptor D1 |
|
| ||
| Canabinoid | ||
|
| ||
| Cnr1 | 2.16 | cannabinoid receptor 1 (brain) |
Expression of genes for neuropeptides and their receptors was also altered (Table 2; Supplementary Table 1). In many cases the expression of the neuropeptide and receptors had parallel changes. Pdyn (prodynorphin) and its receptor Oprk1 were both decreased, as was Pnoc (prepronociceptin) and its receptor Oprl1, galanin and two of its receptors, and Neuromedin B and its receptor. For others, there appeared to be compensatory changes, with the expression of the peptide and its receptors changing in opposite directions. Neuropeptide Y (Npy) was increased (+1.6 fold) but its receptors Npy1r and Npy5r were both decreased (−1.6 fold). There was a 3-fold increase in the gene expression for hypocretin (orexin), but expression of both hypocretin receptors was decreased about 2-fold. Expression of Cck (cholecystokinin) was increased but its B receptor gene was decreased. There was a small decrease in expression of Sst (somatostatin) with decreases in two of its receptors (Sstr1 and Sstr2) and a large increase in a third receptor, Sstr3. The two corticotropin-releasing hormone (Crh) receptors were differentially altered, with Crhr1 increased and Crhr2 decreased.
Table 2. Differentially expressed neuropeptides.
Neuropeptide and receptor genes whose expression is altered by adolescent ethanol binge-drinking (FDR<0.05)
| Gene | Fold Change | Gene Name |
|---|---|---|
| Parallel changes | ||
|
| ||
| Pdyn | −1.5 | Preprodynorphin |
| Oprk1 | −1.4 | Kappa opioid receptor1 |
| Pnoc | −1.3 | Prepronociceptin |
| Oprl1 | −1.5 | Opioid receptor like 1 |
| Gal | −2.1 | Galanin |
| Galr1 | −2 | galanin receptor 1 |
| Galr2 | −1.8 | galanin receptor 2 |
| Nmbr | −2.3 | Neuromedin B |
| Nmbr | −1.7 | Neuromedin B receptor |
| Nts | −1.8 | Neurotensin |
| Ntsr1 | −1.4 | Neurotensin receptor 1 |
|
| ||
| Compensatory changes | ||
|
| ||
| Npy | 1.6 | Neuropeptide Y |
| Npy1r | −1.6 | NPY receptor 1 |
| Npy5r | −1.6 | NPY receptor 5 |
| Hcrt | 3 | hypocretin |
| Hcrtr1 | −2 | hypocretin receptor 1 |
| Hcrtr2 | −2 | hypocretin receptor 2 |
| Cck | 1.5 | Cholecystokinin |
| Cckbr | −1.9 | B receptor for cholecystokinin |
| Sst | −1.3 | Somatostatin |
| Sstr1 | −1.6 | Somatostatin receptor 1 |
| Sstr2 | −1.6 | Somatostatin receptor 2 |
| Sstr3 | 3 | Somatostatin receptor 3 |
|
| ||
| Other | ||
|
| ||
| Oprm1 | −1.6 | Mu opioid receptor |
| Oxtr | −1.6 | Oxytocin receptor |
| Tacr1 | −1.8 | Tachykinin receptor 1 |
| Tacr3 | 1.3 | Tachykinin receptor 3 |
| Nmur2 | −1.8 | Neuromedin U receptor 2 |
| Crhr1 | 3 | corticotropin releasing hormone receptor1 |
| Crhr2 | −1.6 | corticotropin releasing hormone receptor2 |
| Mc4r | −2 | melanocortin receptor 4 |
Expression of many ion channel genes was altered (Table 3): 32 of 67 potassium channels were differentially expressed, with 21 increased and 11 decreased; Kcnj6 was increased 30%. Twelve calcium channels showed altered expression, with equal numbers increased and decreased. Three sodium channels were increased and 6 decreased. Two chloride channel genes were increased and 4 decreased.
Table 3. Differentially expressed Ion Channels.
Ion Channel genes whose expression is altered by adolescent ethanol binge-drinking (FDR< 0.05)
| GENE | Fold Change | Gene Name |
|---|---|---|
| Potassium channels | 34 of 67 identified | |
|
| ||
| Kcna1 | 1.35 | potassium voltage-gated channel, shaker-related subfamily, member |
| Kcna3 | −1.48 | potassium voltage-gated channel, shaker-related subfamily, member 3 |
| Kcna4 | −1.41 | potassium voltage-gated channel, shaker-related subfamily, member 4 |
| Kcna6 | −1.35 | potassium voltage-gated channel, shaker-related subfamily, member 6 |
| Kcnab1 | 1.42 | potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| Kcnab3 | 1.4 | potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| Kcnc1 | 1.51 | potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| Kcnc2 | −1.24 | potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| Kcnc3 | 3.08 | potassium voltage-gated channel, Shaw-related subfamily, member 3 |
| Kcnd2 | 1.63 | potassium voltage-gated channel, Shal-related subfamily, member 2 |
| Kcng1 | 1.79 | potassium voltage-gated channel, subfamily G, member 1 |
| Kcng4 | 2.15 | potassium voltage-gated channel, subfamily G, member 4 |
| Kcnh1 | 2.24 | potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| Kcnh3 | 4.54 | potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| Kcnh5 | −1.4 | potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| Kcnj12 | 2.64 | potassium inwardly-rectifying channel, subfamily J, member 12 |
| Kcnj3 | 1.7 | potassium inwardly-rectifying channel, subfamily J, member 3 |
| Kcnj6 | 1.31 | potassium inwardly-rectifying channel, subfamily J, member 6 |
| Kcnk1 | 1.51 | potassium channel, subfamily K, member 1 |
| Kcnk10 | 1.35 | potassium channel, subfamily K, member 10 |
| Kcnk12 | 3.06 | potassium channel, subfamily K, member 12 |
| Kcnk16 | 2.64 | potassium channel, subfamily K, member 16 |
| Kcnk2 | −1.51 | potassium channel, subfamily K, member 2 |
| Kcnk3 | 1.6 | potassium channel, subfamily K, member 3 |
| Kcnk9 | −1.6 | potassium channel, subfamily K, member 9 |
| Kcnmb4 | 1.44 | potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| Kcnn3 | −1.73 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| Kcnq3 | −1.39 | potassium voltage-gated channel, KQT-like subfamily, member 3 |
| Kcns3 | −1.47 | potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| Kcnt1 | 1.74 | potassium channel, subfamily T, member 1 |
| Kcnt2 | −1.61 | potassium channel, subfamily T, member 2 |
| Kcnv1 | 1.88 | potassium channel, subfamily V, member 1 |
|
| ||
| Sodium Channels | 9 of 14 identified | |
|
| ||
| Scn1b | 2.08 | sodium channel, voltage-gated, type I, beta subunit |
| Scn3a | −1.46 | sodium channel, voltage-gated, type III, alpha subunit |
| Scn3b | −1.63 | sodium channel, voltage-gated, type III, beta subunit |
| Scn4a | −1.67 | sodium channel, voltage-gated, type IV, alpha subunit |
| Scn4b | 1.9 | sodium channel, voltage-gated, type IV, beta subunit |
| Scn5a | −1.63 | sodium channel, voltage-gated, type V, alpha subunit |
| Scn8a | 1.22 | sodium channel, voltage gated, type VIII, alpha subunit |
| Scn9a | −1.91 | sodium channel, voltage-gated, type IX, alpha subunit |
| Scnn1a | −1.45 | sodium channel, non-voltage-gated 1 alpha subunit |
|
| ||
| Calcium Channels | 12 of 21 identified | |
|
| ||
| Cacna1a | 1.53 | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit/ |
| Cacna1b | −1.25 | calcium channel, voltage-dependent, N type, alpha 1B subunit |
| Cacna1d | 1.22 | calcium channel, voltage-dependent, L type, alpha 1D subu |
| Cacna1g | 1.76 | calcium channel, voltage-dependent, T type, alpha 1G subunit |
| Cacna1h | −1.39 | calcium channel, voltage-dependent, T type, alpha 1H subu |
| Cacna1i | 1.49 | calcium channel, voltage-dependent, T type, alpha 1I subunit |
| Cacna2d1 | −1.47 | calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
| Cacnb1 | −1.26 | calcium channel, voltage-dependent, beta 1 subunit |
| Cacnb4 | 1.69 | calcium channel, voltage-dependent, beta 4 subunit |
| Cacng3 | −1.78 | calcium channel, voltage-dependent, gamma subunit 3 |
| Cacng7 | 1.25 | calcium channel, voltage-dependent, gamma subunit 7 |
| Cacng8 | −1.39 | calcium channel, voltage-dependent, gamma subunit 8 |
|
| ||
| Chloride Channels | 6 of 14 identified | |
|
| ||
| Clcn5 | −1.55 | chloride channel, voltage-sensitive 5 |
| Clcn6 | −1.32 | chloride channel, voltage-sensitive 6 |
| Clcnkb | −2.46 | chloride channel, voltage-sensitive Kb |
| Clic1 | 1.61 | chloride intracellular channel 1 |
| Clic2 | −1.43 | chloride intracellular channel 2 |
| Clic6 | 2.93 | chloride intracellular channel 6 |
Many genes involved in myelin production were decreased by 30 to 60% (Supplementary Table 1). Similar decreases in multiple brain regions have been reported multiple times in human post-mortem studies (Liu et al., 2006a; McClintick et al., 2013) and animal models (Alfonso-Loeches et al., 2012).
3.2 Pathway Analysis
Ingenuity pathway analysis (IPA) identified 153 pathways that changed significantly as a result of adolescent ethanol binge-drinking (at FDR ≤ 0.05; Supplementary Table 2; selected subset in Table 4). Neurotransmitter pathways were significantly altered, especially the serotonin pathway (Table 4). Pathways related to cAMP and CREB signaling were also altered, along with pathways involved in axonal guidance. Inflammatory pathways and pathways related to oxidative stress, such as Hif1α and NRF2-mediated stress responses, were altered. Pathways related to neuropeptides Crh and cholecystokinin/gastrin and to several growth factors (Bmp, Igf1, Ngf, Hgf, Pedf, Fgf and Egf) were altered, as were pathways for transcription factors related to PPAR2, RAR, LXR/RXR, and TR/RXR (Table 4). Several genes were found in a large percentage of the significantly altered pathways, including Crebbp, Jun and Fos, MAP kinases, a group of protein kinase A and C genes, and two NFκB subunits (Rela and Nfkb2) (Supplementary Table 3).
Table 4. Selected pathways altered by adolescent ethanol binge-drinking.
Genes with FDR ≤ 0.05 (from Supplementary Table 1) were analyzed using Ingenuity Pathway Analysis; the full list of pathways that were significantly altered (FDR ≤ 0.05) and the genes within each pathway is in Supplementary Table 2. This is a subset selected for putative relevance.
| Ingenuity Canonical Pathways | FDR | 1 Affected in ACB-sh or CEA |
|---|---|---|
| Neurotransmitters | ||
|
| ||
| Serotonin Receptor Signaling | 1.70E-07 | |
| Dopamine Receptor Signaling | 9.30E-04 | |
| GABA Receptor Signaling | 1.70E-03 | |
| Glutamate Receptor Signaling | 1.70E-03 | |
| Dopamine Degradation | 3.10E-02 | |
| α-Adrenergic Signaling | 3.10E-02 | |
|
| ||
| Neuropeptides | ||
|
| ||
| Corticotropin Releasing Hormone Signaling | 7.10E-04 | |
| Cholecystokinin/Gastrin-mediated Signaling | 1.40E-03 | |
|
| ||
| cAMP, Creb related | ||
|
| ||
| Protein Kinase A Signaling | 1.30E-05 | Acb-sh |
| cAMP-mediated signaling | 4.30E-05 | both |
| Gα12/13 Signaling | 4.30E-05 | |
| Dopamine-DARPP32 Feedback in cAMP Signaling | 2.10E-04 | |
| Gαi Signaling | 3.50E-04 | |
| CREB Signaling in Neurons | 9.10E-04 | |
| P2Y Purigenic Receptor Signaling Pathway | 1.10E-03 | |
| Calcium Transport I | 1.90E-03 | |
| Calcium Signaling | 3.20E-03 | |
| Gαq Signaling | 4.40E-03 | |
| Gαs Signaling | 4.70E-02 | |
|
| ||
| Inflammation | ||
|
| ||
| LPS/IL-1 Mediated Inhibition of RXR Function | 4.30E-05 | |
| CXCR4 Signaling | 9.50E-04 | |
| Acute Phase Response Signaling | 1.10E-03 | Acb-sh |
| LPS-stimulated MAPK Signaling | 2.00E-03 | |
| IL-1 Signaling | 2.90E-03 | |
| TNFR1 Signaling | 3.70E-03 | |
| IL-6 Signaling | 5.50E-03 | |
| CCR3 Signaling in Eosinophils | 1.50E-02 | |
| HMGB1 Signaling | 1.90E-02 | both |
| IL-10 Signaling | 2.90E-02 | |
| IL-17 Signaling | 4.70E-02 | |
| IL-2 Signaling | 4.80E-02 | |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages | 2.90E-03 | |
|
| ||
| Axonal Guidance | ||
|
| ||
| Axonal Guidance Signaling | 4.60E-09 | |
| Ephrin Receptor Signaling | 9.10E-03 | |
| Netrin Signaling | 4.70E-02 | |
|
| ||
| Oxidative stress | ||
|
| ||
| NRF2-mediated Oxidative Stress Response | 3.20E-04 | |
| Aryl Hydrocarbon Receptor Signaling | 1.80E-03 | Acb-sh |
| HIF1α Signaling | 6.30E-03 | |
| Angiopoietin Signaling | 1.00E-02 | |
| Erythropoietin Signaling | 2.60E-02 | |
|
| ||
| Growth factors | ||
|
| ||
| BMP signaling pathway | 3.20E-05 | |
| IGF-1 Signaling | 9.10E-04 | Acb-sh |
| NGF Signaling | 2.90E-03 | |
| HGF Signaling | 8.10E-03 | |
| PEDF Signaling | 1.30E-02 | |
| FGF Signaling | 1.50E-02 | |
| EGF Signaling | 2.10E-02 | |
| Neurotrophin/TRK Signaling | 2.90E-02 | |
| TGF-β Signaling | 2.30E-03 | |
|
| ||
| Transcription Factors | ||
|
| ||
| PPARα/RXRα Activation | 1.00E-04 | |
| RAR Activation | 2.10E-04 | Acb-sh |
| PPAR Signaling | 5.50E-04 | |
| LXR/RXR Activation | 1.80E-03 | |
| TR/RXR Activation | 3.90E-02 | |
|
| ||
| Signaling pathways | ||
|
| ||
| Signaling by Rho Family GTPases | 8.70E-05 | |
| Rac Signaling | 1.10E-03 | |
| RhoGDI Signaling | 2.90E-03 | |
| p38 MAPK Signaling | 3.10E-02 | |
| G-Protein Coupled Receptor Signaling | 8.50E-07 | Acb-sh |
Identifies pathways also significantly altered in the accumbens shell or central amydala of these same animals
IPA analysis of upstream regulators provides further evidence for a strong pro-inflammatory response: NFκB and 11 cytokines genes were activated and Il1rn (an antagonist of the IL1 receptor) was decreased (Supplementary Table 4). This analysis also provided additional evidence for increased signaling by CREB and cAMP, and evidence for increased calcium signaling. Growth factors Vegfa, TGFβ1, Bmp4, Kitlg, and Grp were all activated. Stearate biosynthesis and multiple cholesterol pathways, particularly those downstream of Srebf1 and Srebf2, all had decreased expression (Supplementary Table 4).
3.3 Converging evidence
To look for converging evidence from genetic and genomic studies, we compiled a list of genes whose expression is altered in humans, genes implicated by genome wide association studies (GWAS), and genes whose expression was altered in previous studies of alcohol preferring and non-preferring rats (P and NP, including both naïve states and P animals exposed to alcohol). Many genes in the present study overlapped with the previously implicated genes (Figure 2); specific genes are marked in Supplementary Table 1, which has references for all 29 studies used. Since we were interested in how the present data could inform human studies, we examined which pathways were significantly overrepresented from the 713 genes found in the intersection between this study and the human studies, using IPA. 36 pathways were altered (FDR ≤ 0.10, Supplementary Table 5). These include signaling pathways related to serotonin, cAMP, CREB, oxidative stress, dopamine, Crh and axonal guidance. Fos and Jun, which were included in many of the altered pathways, have also been frequently identified in other experiments using P animals and alcohol (Supplementary Table 1). Among the genes in the intersection, 564 were found in the human post-mortem studies even though most examined frontal cortex and none measured the DRN. Among these are several genes related to myelin, which have decreased expression in the DRN as they had in multiple post-mortem human studies (e.g. Mayfield et al., 2002; McClintick et al., 2013).
Figure 2. Overlap of present data with human and other studies using P rats.
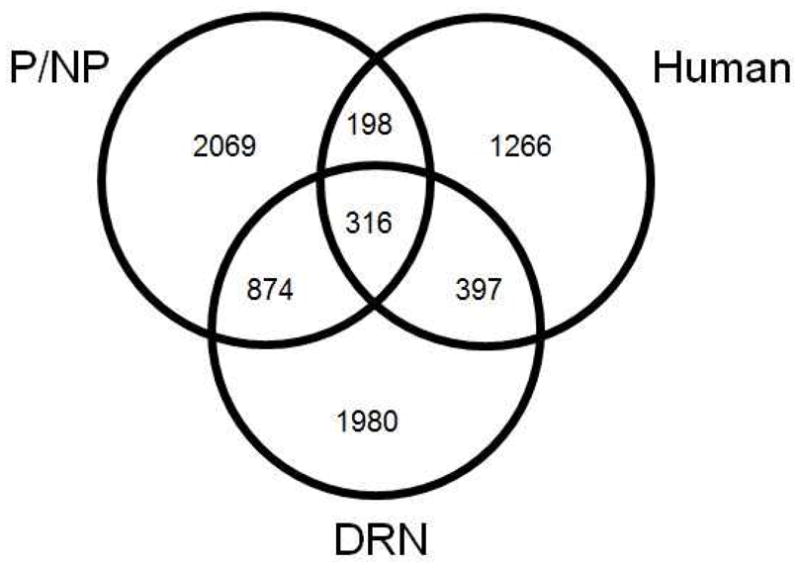
Venn diagram indicating overlap of differentially expressed genes from this study (within the DRN of adolescent binge ethanol-drinking P rats) with previous P/NP animal studies (includes expression studies of naïve P vs. naïve NP and alcohol treated P vs. controls) and human studies (postmortem expression and GWAS). Specific genes and studies are noted in Supplementary Table 1.
4. Discussion
We used RNA sequencing to examine the effect of binge drinking of ethanol during adolescence on gene expression profiles in the dorsal raphe nucleus (DRN), which has not previously been studied in a binge drinking adolescent model. The DRN is a central serotonergic region of the brain (Waselus et al., 2011). It has serotonergic efferents to the reward centers in the brain and is implicated in stress, anxiety and depression (Hale et al., 2012; Waselus et al., 2011). Adolescent P rats consumed 8–10 g ethanol/kg of body weight/day during the three 1-hour sessions, and did this repeatedly for 5 consecutive days during 3 weeks, with a 2-day withdrawal period after each of the initial two weeks (McBride et al., 2014). This repeated binge drinking of ethanol had a profound effect on gene expression in the DRN, with 30% of the identified genes differentially expressed between the ethanol drinking and control animals (Supplementary Table 1). Key functional groups of genes that were altered in expression included neurotransmitter systems (Table 1), neuropeptide systems (Table 2), ion channels (Table 3), and genes related to cAMP, CREB, inflammation, oxidative stress, axonal guidance as well as many growth factors and transcription factors (Table 4, Supplementary Table 2). Pathways identified from genes overlapping with human studies (Supplementary Table 5) highlight pathways related to oxidative stress, inflammation, cAMP, CREB, axonal guidance, dopamine and serotonin.
The DRN is a main origin of the serotonin system, so it is significant that all of the 21 differentially expressed genes in the serotonin pathway are decreased after the repeated binge drinking. This could reflect direct down-regulation of the serotonin system, or the retarded development of the serotonin system. Rodents prenatally exposed to alcohol have decreased serotonin and serotonin metabolism that persists into adulthood resulting in impaired impulse control and changes in other serotonin dependent related behaviors (Macri et al., 2007). If the decrease in serotonin function for these binge drinking adolescents persists into adulthood, these animals could have similar outcomes.
Among the 12 GABAA genes expressed in the DRN, 8 GABAA subunits showed reduced expression. Two subunits, Gabra6 and Gabrd, showed >4-fold increases. Thus, the balance of GABAA subunits was significantly altered, suggesting altered local inhibitory regulation within the DRN. The Gabrd subunit is found in extrasynaptic GABAA receptors composed primarily of α4 or α6 subunits along with β subunits (Lovinger and Roberto, 2013), which generate tonic inhibitory conductance that results in spontaneous low firing rates (Hanchar et al., 2005). These receptors are sensitive to ethanol, which potentiates this tonic current in a protein kinase C delta dependent manner (Lovinger and Roberto, 2013). Prkcd is also increased 2.4 fold. Gad1 (encoding glutamate decarboxylase 1, a rate-limiting enzyme for GABA synthesis) showed significant increases in expression following binge-like drinking; polymorphisms in Gad1 are associated with age of onset for alcohol dependence and initial alcohol sensitivity (Kuo et al., 2009).
NMDA receptors vary in their response to ethanol depending on which subunits make up the receptor, reviewed in (Lovinger and Roberto, 2013). Binge drinking decreased the expression of Grin2b (encoding NR2B; −1.6-fold) and increased Grin2c (encoding NR2C, +2.6-fold) (Table 1). NR2C-containing receptors are less sensitive to inhibition by ethanol than those that contain NR2B (Lovinger and Roberto, 2013), so this shift in ratio should result in decreased sensitivity of the NMDA receptors to ethanol in the DRN, decreasing its inhibitory effects and potentially increasing tolerance to ethanol’s intoxicating effects. Furthermore, the net increased expression of glutamate receptor genes in combination with a net decreased expression of GABAA receptor genes (Table 1) could result in a higher excitatory tone regulating the DRN.
Many neuropeptide systems in the DRN, particularly those related to anxiety and stress (Npy, Crh, galanin, hypocretin/oxytocin – reviewed in Kormos and Gaszner (2013), were altered by binge drinking (Table 2). Npy and Crh work in opposition and must be balanced to maintain homeostasis and resistance to stress (Gilpin, 2012; Sajdyk et al., 2004; Valdez and Koob, 2004). Npy is increased but receptors Npy1r and Npy5r are decreased, perhaps as an adaptation to increased Npy. In contrast to the DRN, Npy5r was found to be increased in both the accumbens shell (Acb-sh) and central nucleus of the amygdala (CeA) of these same animals (McBride et al., 2014). Crh is anxiogenic (Valdez and Koob, 2004). P rats have lower Crh in multiple brain regions, increased Crhr1 receptors (Ehlers et al., 1992), and decreased levels of Crhr2 in several regions (Yong et al., 2014). These two receptors have different affinities for Crh, with Crhr1 binding Crh at low concentrations and Crhr2 binding only at high concentrations. Crhr1 is activated by acute stress and initiates decreased serotonin release and active coping, while Crhr2 is activated by repeated stress and increases serotonin release and passive coping (Valentino, Lucki et al. 2010). In the DRN of these binge drinking adolescent animals, Crhr1 increased 3 fold and Crhr2 decreased 1.6 fold, which could contribute to decreased serotonin levels in these animals.
Galanin mainly has a hyperpolarizing inhibitory effect and is implicated in anxiety, stress, depression and drug addiction, reviewed in Holmes and Picciotto (2006). Galanin and its receptors were decreased about 2 fold indicating a decreased function for this peptide in the DRN. Galanin receptors on serotonin neurons in the DRN can have an inhibitory effect on serotonin signaling (Holmes and Picciotto, 2006). Galr1 is linked to Gi/Go proteins which inhibit adenylyl cyclase. The decrease in Galanin and the receptor may contribute to the overall increase in CREB signaling seen in the DRN of these animals.
Hypocretin is mainly expressed in the lateral hypothalamus. Unexpectedly, the mRNA for hypocretin was detected in the DRN and was 3 fold higher in the binge-drinking rats. The hypocretin mRNA may be in those axons that project from the hypothalamus to innervate the DRN (Xu et al., 2013). Although there is no difference in hypocretin mRNA levels between naïve iP and iNP animals, alcohol drinking increased the hypocretin mRNA levels in iP rats (Lawrence et al., 2006). A hypocretin antagonist (SB-334867) caused a significant decrease in response to ethanol in a cue induced reinstatement experiment (Lawrence et al., 2006). The DRN provides negative feedback via serotonergic projections to the lateral hypothalamus (Xu et al., 2013), so decreased serotonin signaling from the DRN could be expected to lead to decreased inhibition of hypocretin signaling. Both hypocretin receptors (Hcrtr1 and Hcrtr2) were decreased about 2 fold, which could intensify this effect.
There is strong evidence for a neuroinflammatory response, with NFκB and multiple cytokines activated, altering expression of many of their downstream targets including the acute phase response pathway. Chronic alcoholics are known to have highly activated NFκB signaling in the hippocampus (McClintick et al., 2013) and NFκB can be instrumental in the addiction process (Crews et al., 2011). There is evidence for increased oxidative stress which can accompany inflammatory responses (Table 4). There were also indications of an inflammatory response in the Acb-sh of these animals but not the CeA (McBride et al., 2014) with acute phase signaling and the IL-2 pathway altered in Acb-sh. The overlap analysis with the human data (Figure 2, Supplementary Tables 1 & 5) supports previous findings that both oxidative stress and inflammation are important factors in alcohol use disorders in humans.
CREB signaling is increased in the DRN, with the expression of 67 downstream targets altered (Supplementary Table 4). CREB must be phosphorylated to be active and acute alcohol increases phosphorylated CREB (p-CREB) in the CeA and medial amygdala in P but not NP rats (Pandey et al., 2005). Increasing p-CREB in the CeA of P rats, by activating protein kinase A, decreases anxiety and ethanol consumption (Pandey et al., 2005). CREB signaling activates genes necessary for the survival of neurons under stress (Volakakis et al., 2010). CREB can be activated by multiple kinases, several of which are themselves altered, e.g. Camk4 (3.0-fold), Camk2b (1.3-fold), and Protein Kinase A (Prkaca −1.3-fold). Genes induced by CREB include NR4A nuclear receptors (Nr4a1, Nr4a2 and Nr4a3), with expression increased 3- to 6-fold in the binge-drinking adolescents. These receptors are necessary for the increased transcription of some of the neuroprotective genes downstream of CREB (Volakakis et al., 2010). Ppargc1a expression is also increased, and it is responsible for inducing another set of neuroprotective genes that respond to oxidative stress (Volakakis et al., 2010). The increased CREB signaling could in part be responding to oxidative or excitotoxic stress in these animals. The acumbens shell also showed increased CREB signaling in these animals, but the central amygdala did not (McBride et al., 2014). Chronic alcohol has been shown to decrease the phosphorylation of CREB, in other brain regions (Crews and Nixon, 2009). cAMP-mediated signaling was affected in both the acumbens shell and central amygdala of these animals(McBride et al., 2014). The overlap analysis with human data (Supplementary Table 1 & 5, Figure 2) suggests that cAMP and CREB signaling also play an important role in alcohol use disorders in humans.
There were 34 K+ channel subunits that had differential gene expression with a 2 to 1 ratio of increased expression with adolescent binge drinking. A double knockout of Kcnc1 (+1.5) and Kcnc3 (+3.1) in mice is related to hyperactivity and alcohol hypersensitivity (Espinosa et al., 2001). Variants in Kcnj6 (+1.3), an inwardly rectifying potassium channel, were found to be associated with an electrophysiological endophentype related to alcohol dependence (Kang et al., 2012). Kcnh3 had a 4.5 fold increase; a knockout of this gene in mice is associated with hippocampal hyperexciteability and seizures (Zhang et al., 2010). On the other hand, 6 of 9 Na+ channel subunit genes had reduced expression with adolescent binge drinking. Overall, these results suggest changes in neuronal firing and interactions within the DRN that could alter the function of regions receiving serotonin inputs from the DRN.
The repeated binge paradigm had its major impact on the DRN. The CeA and Acb-sh of these same animals showed many fewer genes with altered expression, 309 in the central amygdala and 154 in the Acb-sh (McBride et al., 2014). Sixteen of the 22 altered pathways in the Acb-sh and 3 of the 4 in CeA were also altered in the DRN (noted in Table 4 and Supplementary Table 2). The large difference in numbers of altered genes might be in part attributable to the differences in the cell populations of these 3 regions; the DRN is more focused on the serotonin system, with nearly 50% of the neurons serotonergic (Waselus et al., 2011). More heterogeneity in the other regions could dilute the changes and make them more difficult to identify. Also, differences in the postnatal development of the DRN compared to the other regions may make it more vulnerable to ethanol. Another potential contributor to the difference among the regions is the different technology used to analyze those regions; the RNA sequencing used here may be more sensitive to changes. Technical differences cannot, however, be the primary reason because additional regions also examined by RNA sequencing showed many fewer alterations than the DRN (unpublished data).
5. Conclusion
The DRN is an important region that integrates inputs from many systems, and can have far-reaching effects via the serotonergic projections. Binge drinking in adolescent P rats altered the expression of nearly 30% of the genes in the DRN, suggesting major alterations in DRN functioning. Numerous systems are affected, including multiple neurotransmitter systems, prominent among them the serotonin system (Table 1). Because the DRN has efferents to many brain regions, including the reward systems, the down-regulation of the serotonin system could have far-reaching effects on the brain and its response to alcohol. Neuropeptide systems (Table 2) are also prominently altered, in ways that indicate increased anxiety and decreased tolerance to stress at this time point, 3 h after the last exposure to alcohol. Stress and oxidative damage responses were also greatly affected. The large effects of repeated binges on gene expression in this key region could have major impacts on responses to alcohol and other stressors, which might persist into adulthood.
Supplementary Material
Supplementary Table 1. List of all genes whose expression is altered by binge drinking, at FDR < 0.05. Includes annotation of genes identified by other P/NP studies (differences in the accumbens shell and central amygdala, number of regions with alcohol effects or where P differs from NP, and total number of regions implicated by either naïve or alcohol exposure studies) and by human GWAS and gene expression studies.
Supplementary Table 2. Pathways altered by ethanol exposure Genes with FDR ≤ 0.05 (from Supplementary Table 1) were analyzed using QIAGEN’s Ingenuity® Pathway Analysis; pathways that were significantly altered (FDR ≤ 0.05) are shown here. A selected subset of these is also shown in Table 4.
Supplementary Table 3. Differentially expressed gene found in many pathways.
The 25 genes represented in the most pathways from Supplementary Table 2 are shown, along with their fold change and the number of pathways they appear in.
Supplementary Table 4. Upstream regulators predicted to be activated or repressed, using QIAGEN’s Ingenuity® Upstream analysis. The genes listed in Supplementary Table 1 (altered at FDR ≤ 0.05) were analyzed. Those regulators predicted to be altered (with absolute value of the z-score ≥ 2 are shown.
Supplementary Table 5. Pathways affected in both human studies and expression in DRN. QIAGEN’s Ingenuity® pathway analysis was carried out on the 713 genes that overlap (see Figure 2) between this study and at least one of the human studies (post-mortem gene expression and GWAS studies listed in Supplementary Table 1). Pathways that were altered at FDR ≤ 0.10 are shown, along with the genes that were altered.
Highlights.
Adolescent binge drinking altered gene expression in the rat Dorsal Raphe Nucleus.
Serotonin gene expression in the DRN was decreased by adolescent binge drinking.
Expression of 8 GABA receptor subunits was decreased by adolescent binge drinking.
Neuropeptide systems related to stress/anxiety are dysregulated by binge drinking.
CREB signaling is activated by adolescent binge drinking.
Acknowledgments
Role of Funding source
No commercial external funding, NIH funding only.
This study was funded by grants from the National Institute on Alcohol Abuse and Alcoholism U01AA020892 (HJE), U01AA013522 (RLB) as part of the Integrative Neuroscience Initiative on Alcoholism (INIA-West). The RNA sequencing was done in the Center for Medical Genomics at the Indiana University School of Medicine.
Abbreviations
- Acb-sh
nucleus accumbens shell
- CeA
central nucleus of the amygdala
- CREB
cAMP response element-binding protein
- DRN
dorsal raphe nucleus
- HAD/LAD
High Alcohol and Low Alcohol Drinking rats
- p-CREB
phosphorylated CREB
- P/NP
Alcohol Preferring and Non-Preferring rats
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfonso-Loeches S, Pascual M, Gomez-Pinedo U, Pascual-Lucas M, Renau-Piqueras J, Guerri C. Toll-like receptor 4 participates in the myelin disruptions associated with chronic alcohol abuse. Glia. 2012;60:948–964. doi: 10.1002/glia.22327. [DOI] [PubMed] [Google Scholar]
- Bell RL, Kimpel MW, McClintick JN, Strother WN, Carr LG, Liang T, Rodd ZA, Mayfield RD, Edenberg HJ, McBride WJ. Gene expression changes in the nucleus accumbens of alcohol-preferring rats following chronic ethanol consumption. Pharmacol Biochem Behav. 2009;94:131–147. doi: 10.1016/j.pbb.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Franklin KM, Hauser SR, Engleman EA. Next stop dependence. Binge drinking on the road to alcoholism: preclinical findins on its neurobiology from rat animal models. In: Harris SB, editor. Binge eating and binge drinking: Psychological, social and medical implications. NewYork, NY: Nova Science Publishers; 2013. pp. 1–60. [Google Scholar]
- Bell RL, Rodd ZA, Engleman EA, Toalston JE, McBride WJ. Scheduled access alcohol drinking by alcohol-preferring (P) and high-alcohol-drinking (HAD) rats: modeling adolescent and adult binge-like drinking. Alcohol. 2014;48:225–234. doi: 10.1016/j.alcohol.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Smith RJ, Toalston JE, Franklin KM, McBride WJ. Modeling binge-like ethanol drinking by peri-adolescent and adult P rats. Pharmacology Biochemistry and Behavior. 2011;100:90–97. doi: 10.1016/j.pbb.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Sable HJ, Colombo G, Hyytia P, Rodd ZA, Lumeng L. Animal models for medications development targeting alcohol abuse using selectively bred rat lines: neurobiological and pharmacological validity. Pharmacol Biochem Behav. 2012;103:119–155. doi: 10.1016/j.pbb.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese MR, Liu Y. NGSUtils: a software suite for analyzing and manipulating next-generation sequencing datasets. Bioinformatics (Oxford, England) 2013;29:494–496. doi: 10.1093/bioinformatics/bts731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell AD, Kohl RR, McBride WJ. Serotonin-3 receptor and ethanol-stimulated somatodendritic dopamine release. Alcohol. 1996;13:569–574. doi: 10.1016/s0741-8329(96)00069-9. [DOI] [PubMed] [Google Scholar]
- Campbell AD, McBride WJ. Serotonin-3 receptor and ethanol-stimulated dopamine release in the nucleus accumbens. Pharmacol Biochem Behav. 1995;51:835–842. doi: 10.1016/0091-3057(95)00050-7. [DOI] [PubMed] [Google Scholar]
- Crews FT, Nixon K. Mechanisms of neurodegeneration and regeneration in alcoholism. Alcohol Alcohol. 2009;44:115–127. doi: 10.1093/alcalc/agn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews FT, Zou J, Qin L. Induction of innate immune genes in brain create the neurobiology of addiction. Brain Behav Immun. 2011;25(Suppl 1):S4–S12. doi: 10.1016/j.bbi.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding ZM, Oster SM, Hauser SR, Toalston JE, Bell RL, McBride WJ, Rodd ZA. Synergistic self-administration of ethanol and cocaine directly into the posterior ventral tegmental area: involvement of serotonin-3 receptors. J Pharmacol Exp Ther. 2012;340:202–209. doi: 10.1124/jpet.111.187245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Strother WN, McClintick JN, Tian H, Stephens M, Jerome RE, Lumeng L, Li TK, McBride WJ. Gene expression in the hippocampus of inbred alcohol-preferring and -nonpreferring rats. Genes Brain Behav. 2005;4:20–30. doi: 10.1111/j.1601-183X.2004.00091.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Chaplin RI, Wall TL, Lumeng L, Li TK, Owens MJ, Nemeroff CB. Corticotropin releasing factor (CRF): studies in alcohol preferring and non-preferring rats. Psychopharmacology (Berl) 1992;106:359–364. doi: 10.1007/BF02245418. [DOI] [PubMed] [Google Scholar]
- Engleman EA, Rodd ZA, Bell RL, Murphy JM. The role of 5-HT3 receptors in drug abuse and as a target for pharmacotherapy. CNS & neurological disorders drug targets. 2008;7:454–467. doi: 10.2174/187152708786927886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa F, McMahon A, Chan E, Wang S, Ho CS, Heintz N, Joho RH. Alcohol hypersensitivity, increased locomotion, and spontaneous myoclonus in mice lacking the potassium channels Kv3.1 and Kv3.3. J Neurosci. 2001;21:6657–6665. doi: 10.1523/JNEUROSCI.21-17-06657.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard SG, Duhalde-Vega M, Tagliaferro P, Mirochnic S, Caltana LR, Brusco A. A low chronic ethanol exposure induces morphological changes in the adolescent rat brain that are not fully recovered even after a long abstinence: an immunohistochemical study. Exp Neurol. 2006;200:438–459. doi: 10.1016/j.expneurol.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, Harrison E, Matsumoto I, Wilce PA. Genes associated with alcohol abuse and tobacco smoking in the human nucleus accumbens and ventral tegmental area. Alcohol Clin Exp Res. 2010;34:1291–1302. doi: 10.1111/j.1530-0277.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- Flatscher-Bader T, van der Brug M, Hwang JW, Gochee PA, Matsumoto I, Niwa S, Wilce PA. Alcohol-responsive genes in the frontal cortex and nucleus accumbens of human alcoholics. J Neurochem. 2005;93:359–370. doi: 10.1111/j.1471-4159.2004.03021.x. [DOI] [PubMed] [Google Scholar]
- Gilpin NW. Neuropeptide Y (NPY) in the extended amygdala is recruited during the transition to alcohol dependence. Neuropeptides. 2012;46:253–259. doi: 10.1016/j.npep.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gongwer MA, Murphy JM, McBride WJ, Lumeng L, Li TK. Regional brain contents of serotonin, dopamine and their metabolites in the selectively bred high- and low-alcohol drinking lines of rats. Alcohol. 1989;6:317–320. doi: 10.1016/0741-8329(89)90089-x. [DOI] [PubMed] [Google Scholar]
- Hack LM, Kalsi G, Aliev F, Kuo PH, Prescott CA, Patterson DG, Walsh D, Dick DM, Riley BP, Kendler KS. Limited associations of dopamine system genes with alcohol dependence and related traits in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD) Alcohol Clin Exp Res. 2011;35:376–385. doi: 10.1111/j.1530-0277.2010.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale MW, Shekhar A, Lowry CA. Stress-related serotonergic systems: implications for symptomatology of anxiety and affective disorders. Cellular and molecular neurobiology. 2012;32:695–708. doi: 10.1007/s10571-012-9827-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanchar HJ, Dodson PD, Olsen RW, Otis TS, Wallner M. Alcohol-induced motor impairment caused by increased extrasynaptic GABA(A) receptor activity. Nat Neurosci. 2005;8:339–345. doi: 10.1038/nn1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A, Picciotto MR. Galanin: a novel therapeutic target for depression, anxiety disorders and drug addiction? CNS & neurological disorders drug targets. 2006;5:225–232. doi: 10.2174/187152706776359600. [DOI] [PubMed] [Google Scholar]
- Homer N, Merriman B, Nelson SF. BFAST: an alignment tool for large scale genome resequencing. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Yamamoto M, Ozawa H, Saito T, Kato T. Decreased expression of NEFH and PCP4/PEP19 in the prefrontal cortex of alcoholics. Neurosci Res. 2004;49:379–385. doi: 10.1016/j.neures.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Johnson C, Drgon T, Walther D, Uhl GR. Genomic regions identified by overlapping clusters of nominally-positive SNPs from genome-wide studies of alcohol and illegal substance dependence. PLoS One. 2011;6:e19210. doi: 10.1371/journal.pone.0019210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan L, Wang G, Radovich M, Schneider BP, Clare SE, Wang Y, Liu Y. Potential roles of microRNAs in regulating long intergenic noncoding RNAs. BMC medical genomics. 2013;6(Suppl 1):S7. doi: 10.1186/1755-8794-6-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Almasy L, Brooks A, Chorlian DB, Dick D, et al. Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes Brain Behav. 2012;11:712–719. doi: 10.1111/j.1601-183X.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin Exp Res. 2011;35:963–975. doi: 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol. 2007;41:95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF. Addiction is a Reward Deficit and Stress Surfeit Disorder. Frontiers in psychiatry. 2013;4:72. doi: 10.3389/fpsyt.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormos V, Gaszner B. Role of neuropeptides in anxiety, stress, and depression: from animals to humans. Neuropeptides. 2013;47:401–419. doi: 10.1016/j.npep.2013.10.014. [DOI] [PubMed] [Google Scholar]
- Kryger R, Wilce PA. The effects of alcoholism on the human basolateral amygdala. Neuroscience. 2010;167:361–371. doi: 10.1016/j.neuroscience.2010.01.061. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Kalsi G, Prescott CA, Hodgkinson CA, Goldman D, Alexander J, van den Oord EJ, Chen X, Sullivan PF, Patterson DG, et al. Associations of glutamate decarboxylase genes with initial sensitivity and age-at-onset of alcohol dependence in the Irish Affected Sib Pair Study of Alcohol Dependence. Drug Alcohol Depend. 2009;101:80–87. doi: 10.1016/j.drugalcdep.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankford MF, Bjork AK, Myers RD. Differential efficacy of serotonergic drugs FG5974, FG5893, and amperozide in reducing alcohol drinking in P rats. Alcohol. 1996;13:399–404. doi: 10.1016/0741-8329(96)00061-4. [DOI] [PubMed] [Google Scholar]
- Lankford MF, Myers RD. Opiate and 5-HT2A receptors in alcohol drinking: preference in HAD rats is inhibited by combination treatment with naltrexone and amperozide. Alcohol. 1996;13:53–57. doi: 10.1016/0741-8329(95)02011-x. [DOI] [PubMed] [Google Scholar]
- Lawrence AJ, Cowen MS, Yang HJ, Chen F, Oldfield B. The orexin system regulates alcohol-seeking in rats. British journal of pharmacology. 2006;148:752–759. doi: 10.1038/sj.bjp.0706789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24:1873–1882. [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, de Moor MH, Smit AB, Hottenga JJ, Richter MM, Heath AC, et al. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin research and human genetics : the official journal of the International Society for Twin Studies. 2010;13:10–29. doi: 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Iyer VR, Dodd PR, Randall PK, Mayfield RD. Patterns of gene expression in the frontal cortex discriminate alcoholic from nonalcoholic individuals. Neuropsychopharmacology. 2006a;31:1574–1582. doi: 10.1038/sj.npp.1300947. [DOI] [PubMed] [Google Scholar]
- Liu W, Thielen RJ, Rodd ZA, McBride WJ. Activation of serotonin-3 receptors increases dopamine release within the ventral tegmental area of Wistar and alcohol-preferring (P) rats. Alcohol. 2006b;40:167–176. doi: 10.1016/j.alcohol.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lewohl JM, Harris RA, Dodd PR, Mayfield RD. Altered gene expression profiles in the frontal cortex of cirrhotic alcoholics. Alcohol Clin Exp Res. 2007;31:1460–1466. doi: 10.1111/j.1530-0277.2007.00444.x. [DOI] [PubMed] [Google Scholar]
- Long TA, Kalmus GW, Bjork A, Myers RD. Alcohol intake in high alcohol drinking (HAD) rats is suppressed by FG5865, a novel 5-HT1A agonist/5-HT2 antagonist. Pharmacol Biochem Behav. 1996;53:33–40. doi: 10.1016/0091-3057(95)00195-6. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, Roberto M. Behavioral Neurobiology of Alcohol Addiction. Berlin Heidlelberg: Springer; 2013. Synaptic Effects Induced by Alcohol; pp. 31–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macri S, Spinelli S, Adriani W, Dee Higley J, Laviola G. Early adversity and alcohol availability persistently modify serotonin and hypothalamic-pituitary-adrenal-axis metabolism and related behavior: what experimental research on rodents and primates can tell us. Neuroscience and biobehavioral reviews. 2007;31:172–180. doi: 10.1016/j.neubiorev.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Marshall EJ. Adolescent alcohol use: risks and consequences. Alcohol Alcohol. 2014;49:160–164. doi: 10.1093/alcalc/agt180. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Lewohl JM, Dodd PR, Herlihy A, Liu J, Harris RA. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–813. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- McBride WJ. Central nucleus of the amygdala and the effects of alcohol and alcohol-drinking behavior in rodents. Pharmacol Biochem Behav. 2002;71:509–515. doi: 10.1016/s0091-3057(01)00680-3. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Chernet E, Russell RN, Chamberlain JK, Lumeng L, Li TK. Regional CNS densities of serotonin and dopamine receptors in high alcohol-drinking (HAD) and low alcohol-drinking (LAD) rats. Alcohol. 1997a;14:603–609. doi: 10.1016/s0741-8329(97)00072-4. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Chernet E, Russell RN, Wong DT, Guan XM, Lumeng L, Li TK. Regional CNS densities of monoamine receptors in alcohol-naive alcohol-preferring P and -nonpreferring NP rats. Alcohol. 1997b;14:141–148. doi: 10.1016/s0741-8329(96)00117-6. [DOI] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Edenberg HJ, Liang T, Rodd ZA, Bell RL. Changes in gene expression within the extended amygdala following binge-like alcohol drinking by adolescent alcohol-preferring (P) rats. Pharmacol Biochem Behav. 2014;117:52–60. doi: 10.1016/j.pbb.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hauser SR, Edenberg HJ, Bell RL, Rodd ZA. Changes in gene expression within the ventral tegmental area following repeated excessive binge-like alcohol drinking by alcohol-preferring (P) rats. Alcohol. 2013a;47:367–380. doi: 10.1016/j.alcohol.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hyytia P, Colombo G, Edenberg HJ, Lumeng L, Bell RL. Gene expression in the ventral tegmental area of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Pharmacol Biochem Behav. 2012;102:275–285. doi: 10.1016/j.pbb.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hyytia P, Colombo G, Liang T, Edenberg HJ, Lumeng L, Bell RL. Gene expression within the extended amygdala of 5 pairs of rat lines selectively bred for high or low ethanol consumption. Alcohol. 2013b;47:517–529. doi: 10.1016/j.alcohol.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Kimpel MW, Schultz JA, McClintick JN, Edenberg HJ, Bell RL. Changes in gene expression in regions of the extended amygdala of alcohol-preferring rats after binge-like alcohol drinking. Alcohol. 2010;44:171–183. doi: 10.1016/j.alcohol.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Li TK. Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. Critical reviews in neurobiology. 1998;12:339–369. doi: 10.1615/critrevneurobiol.v12.i4.40. [DOI] [PubMed] [Google Scholar]
- McClintick JN, Xuei X, Tischfield JA, Goate A, Foroud T, Wetherill L, Ehringer MA, Edenberg HJ. Stress-response pathways are altered in the hippocampus of chronic alcoholics. Alcohol. 2013;47:505–515. doi: 10.1016/j.alcohol.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, McBride WJ, Lumeng L, Li TK. Contents of monoamines in forebrain regions of alcohol-preferring (P) and -nonpreferring (NP) lines of rats. Pharmacol Biochem Behav. 1987;26:389–392. doi: 10.1016/0091-3057(87)90134-1. [DOI] [PubMed] [Google Scholar]
- NIAAA. NIAAA Council approves definition of Binge Drinking. NIAAA Newsletter. 2004;3:3. [Google Scholar]
- NIAAA. Underage Drinking Fact Sheet 2013 [Google Scholar]
- Overstreet DH, McArthur RA, Rezvani AH, Post C. Selective inhibition of alcohol intake in diverse alcohol-preferring rat strains by the 5-HT2A antagonists amperozide and FG 5974. Alcohol Clin Exp Res. 1997;21:1448–1454. [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Roy A, Xu T. Deficits in amygdaloid cAMP-responsive element-binding protein signaling play a role in genetic predisposition to anxiety and alcoholism. The Journal of clinical investigation. 2005;115:2762–2773. doi: 10.1172/JCI24381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Blanco AM, Cauli O, Miñarro J, Guerri C. Intermittent ethanol exposure induces inflammatory brain damage and causes long-term behavioural alterations in adolescent rats. European Journal of Neuroscience. 2007;25:541–550. doi: 10.1111/j.1460-9568.2006.05298.x. [DOI] [PubMed] [Google Scholar]
- Pascual M, Pla A, Minarro J, Guerri C. Neuroimmune activation and myelin changes in adolescent rats exposed to high-dose alcohol and associated cognitive dysfunction: a review with reference to human adolescent drinking. Alcohol Alcohol. 2014;49:187–192. doi: 10.1093/alcalc/agt164. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 4. San Diego, CA 92101: Academic Press; 1998. [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd-Henricks ZA, McKinzie DL, Edmundson VE, Dagon CL, Murphy JM, McBride WJ, Lumeng L, Li TK. Effects of 5-HT(3) receptor antagonists on daily alcohol intake under acquisition, maintenance, and relapse conditions in alcohol-preferring (P) rats. Alcohol. 2000;21:73–85. doi: 10.1016/s0741-8329(00)00083-5. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, Oster SM, Toalston JE, Pommer TJ, McBride WJ, Murphy JM. Serotonin-3 receptors in the posterior ventral tegmental area regulate ethanol self-administration of alcohol-preferring (P) rats. Alcohol. 2010;44:245–255. doi: 10.1016/j.alcohol.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Gryszowka VE, Toalston JE, Oster SM, Ji D, Bell RL, McBride WJ. The reinforcing actions of a serotonin-3 receptor agonist within the ventral tegmental area: evidence for subregional and genetic differences and involvement of dopamine neurons. J Pharmacol Exp Ther. 2007;321:1003–1012. doi: 10.1124/jpet.106.112607. [DOI] [PubMed] [Google Scholar]
- Sajdyk TJ, Shekhar A, Gehlert DR. Interactions between NPY and CRF in the amygdala to regulate emotionality. Neuropeptides. 2004;38:225–234. doi: 10.1016/j.npep.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Sari Y, Johnson VR, Weedman JM. Role of the serotonergic system in alcohol dependence: from animal models to clinics. Progress in molecular biology and translational science. 2011;98:401–443. doi: 10.1016/B978-0-12-385506-0.00010-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov BP, Jiang L, Trivedi NS, Aston C. Transcription profiling reveals mitochondrial, ubiquitin and signaling systems abnormalities in postmortem brains from subjects with a history of alcohol abuse or dependence. J Neurosci Res. 2003;72:756–767. doi: 10.1002/jnr.10631. [DOI] [PubMed] [Google Scholar]
- Spear L. The Behavioral Neuroscience of Adolescence. New York, NY 10110: W. W. Norten; 2010. [Google Scholar]
- Todd AG, Lin H, Ebert AD, Liu Y, Androphy EJ. COPI transport complexes bind to specific RNAs in neuronal cells. Hum Mol Genet. 2013;22:729–736. doi: 10.1093/hmg/dds480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. 2009 doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 66, 773–784. Bioinformatics (Oxford, England) 2009;25:1105–1111. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood MD, Mann JJ, Arango V. Morphometry of dorsal raphe nucleus serotonergic neurons in alcoholism. Alcohol Clin Exp Res. 2007;31:837–845. doi: 10.1111/j.1530-0277.2007.00365.x. [DOI] [PubMed] [Google Scholar]
- Valdez GR, Koob GF. Allostasis and dysregulation of corticotropin-releasing factor and neuropeptide Y systems: implications for the development of alcoholism. Pharmacol Biochem Behav. 2004;79:671–689. doi: 10.1016/j.pbb.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Valentino RJ, Lucki I, Van Bockstaele E. Corticotropin-releasing factor in the dorsal raphe nucleus: Linking stress coping and addiction. Brain Res. 2010;1314:29–37. doi: 10.1016/j.brainres.2009.09.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volakakis N, Kadkhodaei B, Joodmardi E, Wallis K, Panman L, Silvaggi J, Spiegelman BM, Perlmann T. NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc Natl Acad Sci U S A. 2010;107:12317–12322. doi: 10.1073/pnas.1007088107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, Harari O, Koller DL, Wetherill L, Agrawal A, et al. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry. 2013;18:1218–1224. doi: 10.1038/mp.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RJ, Lallemand F, de Witte P. Influence of adolescent heavy session drinking on the systemic and brain innate immune system. Alcohol Alcohol. 2014;49:193–197. doi: 10.1093/alcalc/agu002. [DOI] [PubMed] [Google Scholar]
- Waselus M, Valentino RJ, Van Bockstaele EJ. Collateralized dorsal raphe nucleus projections: a mechanism for the integration of diverse functions during stress. Journal of chemical neuroanatomy. 2011;41:266–280. doi: 10.1016/j.jchemneu.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak KM, Pert A, Linnoila M. Antagonism of 5-HT3 receptors attenuates the effects of ethanol on extracellular dopamine. European journal of pharmacology. 1990;187:287–289. doi: 10.1016/0014-2999(90)90015-x. [DOI] [PubMed] [Google Scholar]
- Xu TR, Yang Y, Ward R, Gao L, Liu Y. Orexin receptors: multi-functional therapeutic targets for sleeping disorders, eating disorders, drug addiction, cancers and other physiological disorders. Cellular signalling. 2013;25:2413–2423. doi: 10.1016/j.cellsig.2013.07.025. [DOI] [PubMed] [Google Scholar]
- Yong W, Spence JP, Eskay R, Fitz SD, Damadzic R, Lai D, Foroud T, Carr LG, Shekhar A, Chester JA, et al. Alcohol-Preferring Rats Show Decreased Corticotropin-Releasing Hormone-2 Receptor Expression and Differences in HPA Activation Compared to Alcohol-Nonpreferring Rats. Alcohol Clin Exp Res. 2014;38:1275–1283. doi: 10.1111/acer.12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bertaso F, Yoo JW, Baumgartel K, Clancy SM, Lee V, Cienfuegos C, Wilmot C, Avis J, Hunyh T, et al. Deletion of the potassium channel Kv12.2 causes hippocampal hyperexcitability and epilepsy. Nat Neurosci. 2010;13:1056–1058. doi: 10.1038/nn.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Pu CF, Murphy J, Lumeng L, Li TK. Serotonergic neurons in the alcohol preferring rats. Alcohol. 1994;11:397–403. doi: 10.1016/0741-8329(94)90024-8. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci U S A. 2011;108:6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Bierut LJ, Goate A, Hesselbrock V, Kuperman S, et al. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:44–58. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang F, Zhang H, Zhang XY, Wang F, Li CS, Lu L, Hong J, Lu L, Krystal J, et al. Genome-wide search for replicable risk gene regions in alcohol and nicotine co-dependence. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:437–444. doi: 10.1002/ajmg.b.32047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. List of all genes whose expression is altered by binge drinking, at FDR < 0.05. Includes annotation of genes identified by other P/NP studies (differences in the accumbens shell and central amygdala, number of regions with alcohol effects or where P differs from NP, and total number of regions implicated by either naïve or alcohol exposure studies) and by human GWAS and gene expression studies.
Supplementary Table 2. Pathways altered by ethanol exposure Genes with FDR ≤ 0.05 (from Supplementary Table 1) were analyzed using QIAGEN’s Ingenuity® Pathway Analysis; pathways that were significantly altered (FDR ≤ 0.05) are shown here. A selected subset of these is also shown in Table 4.
Supplementary Table 3. Differentially expressed gene found in many pathways.
The 25 genes represented in the most pathways from Supplementary Table 2 are shown, along with their fold change and the number of pathways they appear in.
Supplementary Table 4. Upstream regulators predicted to be activated or repressed, using QIAGEN’s Ingenuity® Upstream analysis. The genes listed in Supplementary Table 1 (altered at FDR ≤ 0.05) were analyzed. Those regulators predicted to be altered (with absolute value of the z-score ≥ 2 are shown.
Supplementary Table 5. Pathways affected in both human studies and expression in DRN. QIAGEN’s Ingenuity® pathway analysis was carried out on the 713 genes that overlap (see Figure 2) between this study and at least one of the human studies (post-mortem gene expression and GWAS studies listed in Supplementary Table 1). Pathways that were altered at FDR ≤ 0.10 are shown, along with the genes that were altered.


