Fig. 2.
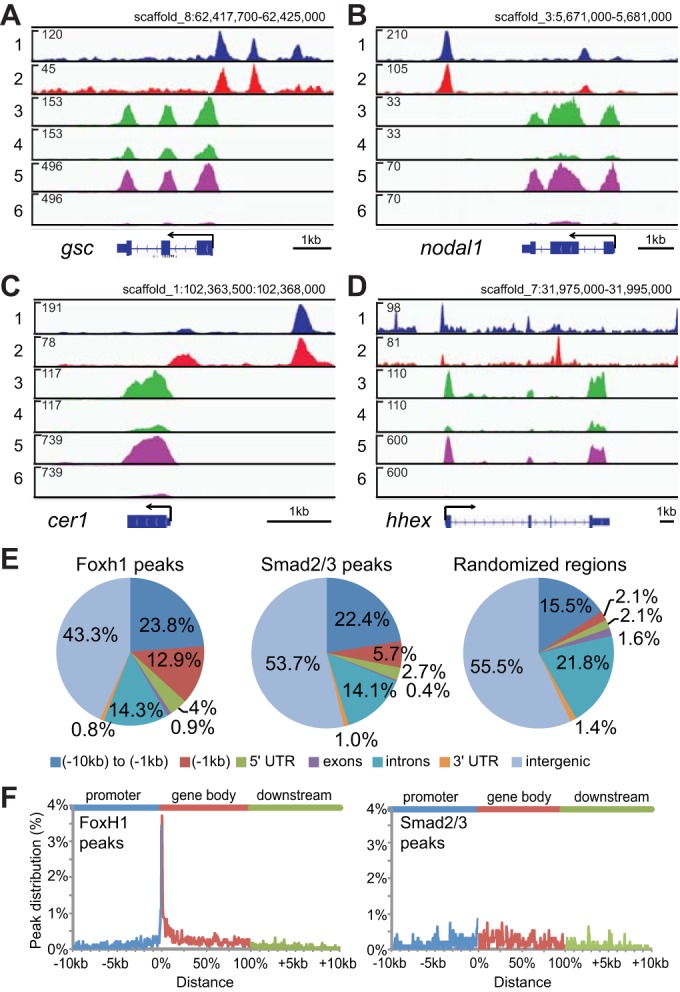
Genome-wide survey of Nodal and Foxh1 targets in early gastrulae. (A-D) IGV genome browser views of gsc (A), nodal1 (B), cer1 (C) and hhex (D) genes. Track contents: (1) Foxh1 ChIP-seq; (2) Smad2/3 ChIP-seq; (3) RNA-seq of uninjected control embryos; (4) RNA-seq of foxh1 MO-injected embryos; (5) RNA-seq of mock-treated control embryos; and (6) RNA-seq of SB431542-treated embryos. The numbers in the upper left of each track indicate track heights. (E) Genome-wide analyses of Foxh1 and Smad2/3 peaks. Pie chart distributions of Foxh1 and Smad2/3 peaks across seven defined genomic features are shown. Randomized regions were also analyzed for comparison (right). These were generated by randomly redistributing the intervals of Foxh1 peaks throughout the genome. In parallel, Smad2/3 peak intervals were similarly randomized and showed a nearly identical genomic distribution (data not shown). (F) Distribution of Foxh1 (left) and Smad2/3 (middle) peaks within the intervals of 10 kb upstream of gene 5′ ends, gene bodies and 10 kb downstream of gene 3′ ends. Supplementary material Fig. S7 contains a distribution of randomly selected regions. As individual gene bodies are highly variable in length, we normalized these segments on a 0-100% scale (Zhang et al., 2012).
