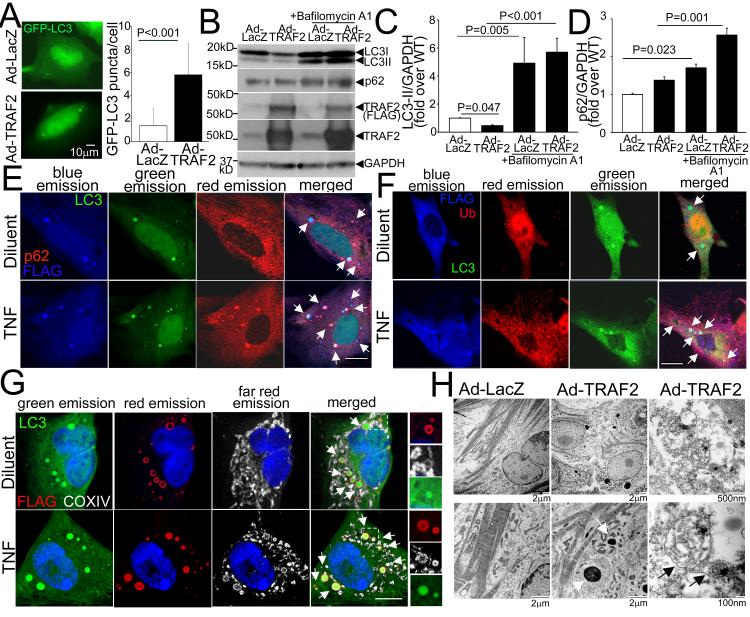
Figure 2. Exogenous TRAF2 localizes to the mitochondria in autophagosomes.
A) Representative immunofluorescence images demonstrating GFP-LC3 expression (left) with quantitation (right) of punctate GFP-LC3 in NRCMs adenovirally transduced with FLAG-TRAF2 or LacZ as control (multiplicity of infection, MOI=100, 48hrs). N=45-52 nuclei/group. P value is by Mann-Whitney test. B-D) Representative immunoblot (B) and quantitation of LC3-II (C) and p62 (D) expression (with GAPDH as loading control) in NRCMs adenovirally transduced with TRAF2 or LacZ (MOI=100, 48hrs) and treated with Bafilomycin A1 (100nmol/l, 24hrs) to assess autophagic flux. N=7/group. P values are by post-hoc pairwise comparisons after Kruskal-Wallis test in C and D. E-G) Representative confocal images demonstrating co-localization of adenovirally-transduced FLAG-tagged TRAF2 with GFP-LC3 and p62 (arrows, in E); with GFP-LC3 and ubiquitin (arrows, in F); and with GFP-LC3 and mitochondria (identified by COX IV immunostaining; arrows, in G) in NRCMs treated with TNF (200ng/ml) or diluent. Boxed area is shown with magnification of individual channels to demonstrate co-localization. Scale bar =10μm. H) Representative transmission electron micrographs of NRCMs adenovirally transduced with TRAF2 or LacZ as control (MOI=100) demonstrating increased autophagic structures enclosing mitochondrial remnants, with TRAF2 overexpression. White arrows point to amorphous proteinaceous deposits; and black arrows point to autophagic structures.