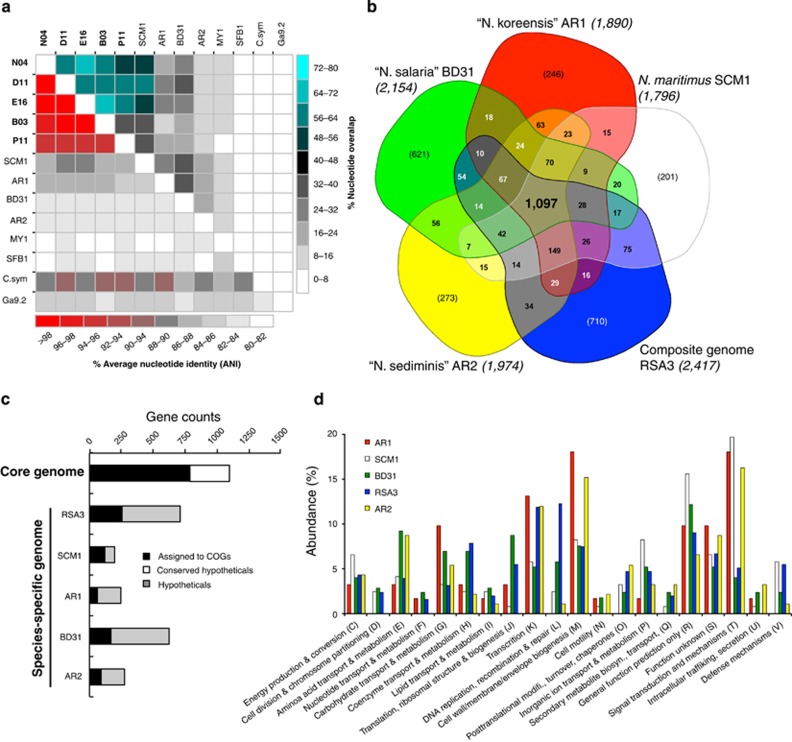
Figure 2.
Comparison of all Nitrosopumilus genomes based on (a) whole-genome alignment of the assembled bases, (b) reciprocal blast analysis of their predicted protein-coding genes, (c) functional assignment of cluster of orthologous genes (COGs) in the core and species-specific genomes and (d) relative abundance of COG categories in their variable genomes. (a) Highlights the relatedness of our SAGs (in bold) to each other than to published genomes, whereas b–d inventories differences in the proteome of the co-assembled composite Nitrosopumilus-like genome RSA3 to four publicly available Nitrosopumilus genomes. (b) The Nitrosopumilus core genome with 1097 genes (in bold), the species-specific gene sets (in bracket) and the flexible genes present in some but not all genomes. The total predicted protein-coding genes in each genome are indicated in brackets next to their names.