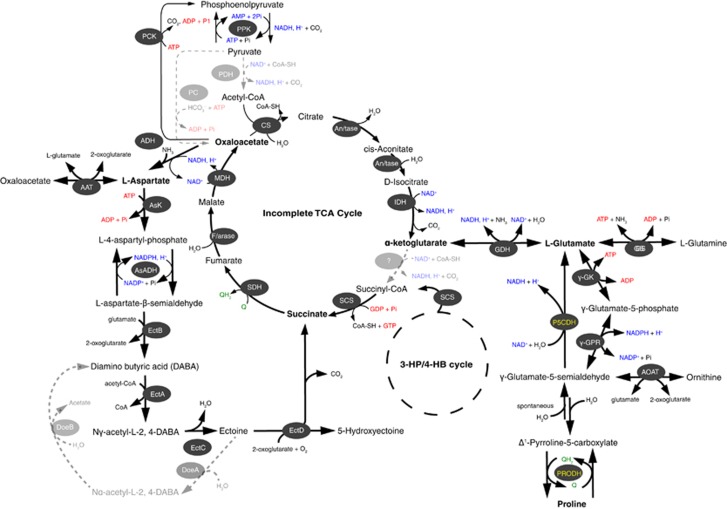
Figure 5.
Metabolic scheme of ectoine, glutamate and proline synthesis and catabolism inferred from the Nitrosopumilus genomes. Enzymes for each pathway are represented as oval shapes and color-coded according to their occurrence in all species (white font), or only in RSA3 and AR2 (yellow font). Gray dashed arrows highlight pathways that are absent in all Nitrosopumilus species. AAT, aspartate aminotransferase; ADH, aspartate dehydrogenase; AOAT, acetylornithine aminotransferase; AsK, aspartate kinase; AsADH, aspartyl-semialdehyde dehydrogenase; GDH, glutamate dehydrogenase; GS, glutamine synthetase; γ-GK, acetylyglutamate kinase; γ-GPR, N-acetyl-γ-glutamyl phosphate reductase; EctA, L-2,4-diaminobutyrate acetyltransferase; EctB, L-2,4-diaminobutyrate transaminase; EctC, ectoine synthase; EctD, ectoine hydrolase; DoeA, ectoine hydrolase; DoeB, Nα-acetly-L-2,4-diaminobutyrate deacetylase; P5CDH, 1-pyrroline-5-carboxylate dehydrogenase; PRODH, proline dehydrogenase. Other enzymes shown are for pyruvate (de)carboxylation, the Tri-carboxylic acid (TCA) cycle and 3-hydroxypropionate/4-hydroxybutyrate (3-HP/4-HB) cycle including: An/tase, aconitase; CS, citrate synthase; F/arase, Fumarase; IDH, isocitrate dehydrogenase; MDH, malate dehydrogenase; PPK, phosphoenolpyruvate (PEP) kinase; PCK, PEP carboxykinase; PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase; SCS, succinyl-CoA synthetase; SDH, succinate dehydrogenase.