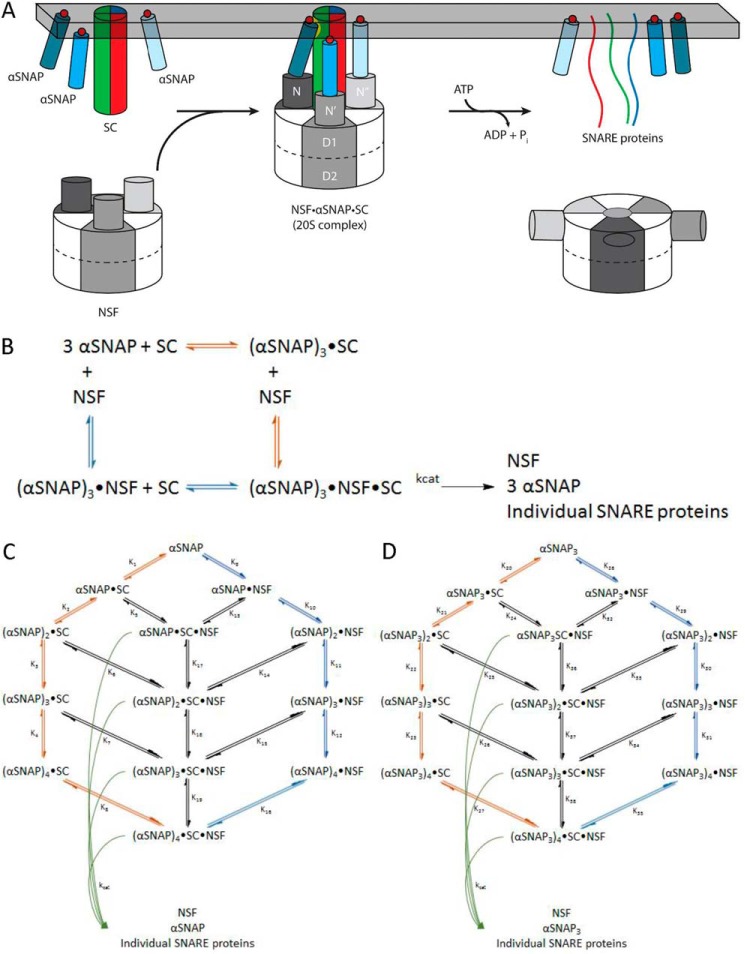
FIGURE 1.
NSF uses the energy of ATP hydrolysis and the αSNAP adaptor to disassemble the SNARE complex. A, NSF binds to the membrane-bound SC via αSNAP, which is probably membrane-associated. Using the energy from ATP hydrolysis, the SC is disassembled into its component proteins, and NSF and αSNAP are available for subsequent rounds of disassembly. B, simplified model for the dissociation of the SC by NSF bound to 3 αSNAP, in which either αSNAP binds SC before NSF (red pathway) or SC binds to αSNAP·NSF to form the 20 S complex (blue pathway in B and C). C, kinetic modeling scheme for the association of n αSNAP molecules with SC and disassembly of the SC by NSF. The red branch represents binding of n αSNAP molecules to the SC to form the complete NSF substrate before interaction with NSF; the blue branch corresponds to n αSNAP molecules binding to NSF before association with the SC. Potential intermediate, non-productive assemblies lacking sufficient αSNAP molecules to support SC disassembly are also shown. For a given model that uses n αSNAP molecules, the 20 S complex will be (αSNAP)n·SC·NSF, which disassembles into n αSNAPs, the individual SNARE proteins, and free NSF (curved green arrows). D, kinetic modeling scheme for the association of n αSNAP3 molecules with SC and disassembly of the SC by NSF. The red branch represents binding of n αSNAP3 molecules to the SC to form the complete NSF substrate before interaction with NSF; the blue branch corresponds to n αSNAP3 molecules binding to NSF before association with the SC. Potential intermediate, non-productive assemblies lacking sufficient αSNAP3 molecules to support SC disassembly are also shown. For a given model that uses n αSNAP3 molecules, the 20 S complex will be (αSNAP3)n·SC·NSF, which disassembles into n αSNAP3s, the individual SNARE proteins, and free NSF (curved green arrows).