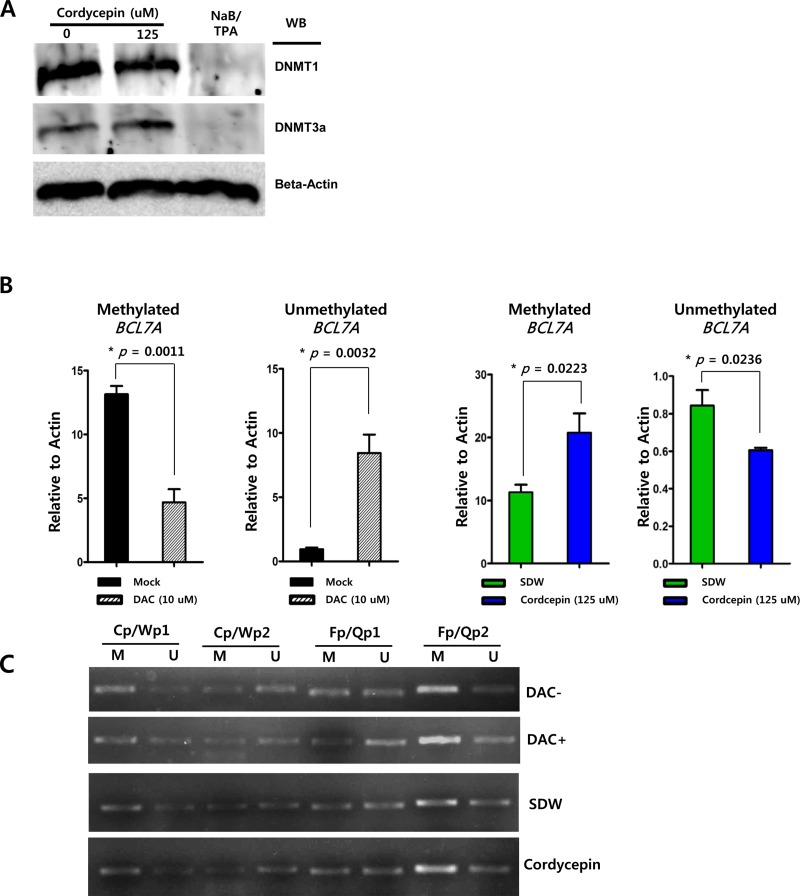
Figure 5. Effect of cordycepin on methylation.
The effect of cordycepin on methylation was determined using Western blot analysis as well as methylation-specific PCR and RT-qPCR assays. (A) Western blot analysis was performed to determine if cordycepin affected DNMT1 and DNMT3a production in SNU719 cells treated with cordycepin. Beta-actin was used as loading and internal controls. These Western blot analysis showed that cordycepin did not affect DNMT1 levels, but did significantly induced DMNT3 expression. (B) Methylation-specific PCR was performed on cordycepin-treated (125 μM) SNU719 cells to determine if cordycepin affected methylation of BCL7A, a tumor suppressor gene. Compared to the mock treatment, 10 μM 5-aza-2′-deoxycytidine (DAC) treatment suppressed methylation by up to 35% and induced demethylation by up to 882% (left two panels). The methylation-specific PCR assay showed that cordycepin treatment (125 μM) induced BCL7A methylation by up to 58% and suppressed demethylation by up to 37% in SUN719 cells (right two panels). P-values <0.05 (95% confidence) were considered statistically significant. (C) An RT-PCR assay was performed to determine if cordycepin affected methylation of EBV genomic loci in SNU719 cells treated with cordycepin (125 μM). Methylation of EBV genomic loci near Cp/Wp promoters was not affected by cordycepin treatment, whereas that around Fp/Qp promoters was enhanced. Specifically, regions downstream of the Fp/Qp promoters were highly methylated in response to cordycepin treatment. Cp/Wp1 and Cp/Wp2 are EBV genomic loci upstream and downstream of Cp/Wp promoters, respectively. Fp/Qp1 and Fp/Qp2 are EBV genomic loci upstream and downstream of Fp/Qp promoters, respectively. M, methylation-specific primers; U, demethylation-specific primers.