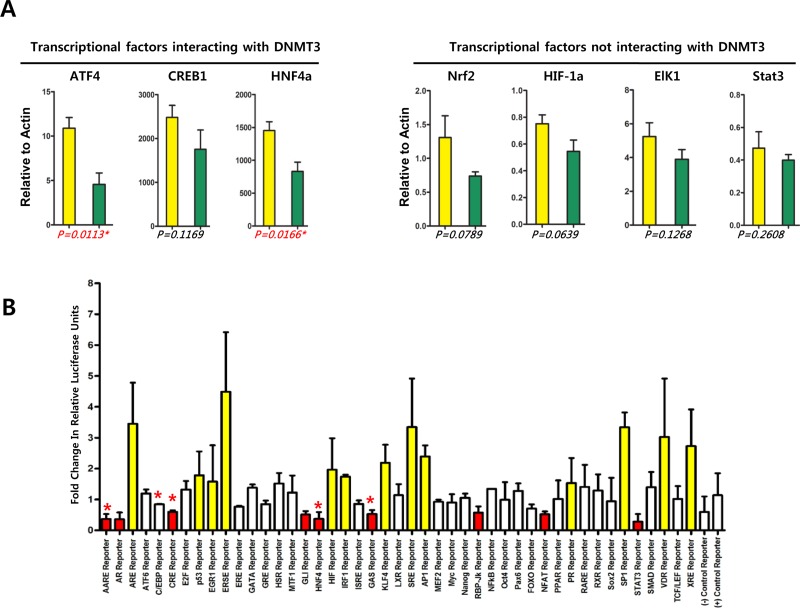
Figure 6. Effect of cordycepin on signal transduction.
A Cignal Finder reporter assay was performed to determine whether cordycepin-induced hypermethylation affects the expression of transcription factors that are known to interact with DNA methyltransferase (DNMT). An RT-qPCR assay was then performed to confirm the expression patterns of transcription factors that were upregulated or downregulated in the Cignal Finder reporter assay. (A) SNU719 cells were treated with cordycepin (125 μM) for 48 h and analyzed using an RT-qPCR assay. Transcription factors kwown not to interact with DNMT3 are Nrf2 (ARE reporter, antioxidant response pathway), YY1 (ERSE reporter, ER stress pathway), HIF-1a (HIF reporter, hypoxia pathway), and Elk1 (SRE reporter, MAPK/EK pathway) [37]. These factors are downregulated by cordycepin, but this effect was not statistically significant. In contrast, transcription factors known to interact with DNMT3 are ATF4 (AARE reporter, amino acid deprivation pathway), CREB1 (CRE reporter, cAMP/PKA pathway) and HNF4 (HNF4 reporter, HNF4 pathway) [37]. These factors are downregulated by cordycepin with statistical significance, especially the expression of ATF4 and HNF4. P-values are listed at the bottom of the graph. (B) SNU719 cells were treated with cordycepin (125 μM) for 48 h and analyzed using the Cignal Finder reporter assay. Reporters induced by cordycepin are shown in yellow, and those suppressed by cordycepin are shown in red. AARE, AR, CRE, GLI, HNF4, GAS, RBP-jK, NFAT, and STAT3 reporters were downregulated by cordycepin. ARE, p53, EGR1, ERSE, HIF, IRF1, KLF4, SRE, AP1, PR, SP1, VDR, and XRE reporters were upregulated by cordycepin. Each reporter assay was performed in triplicate.