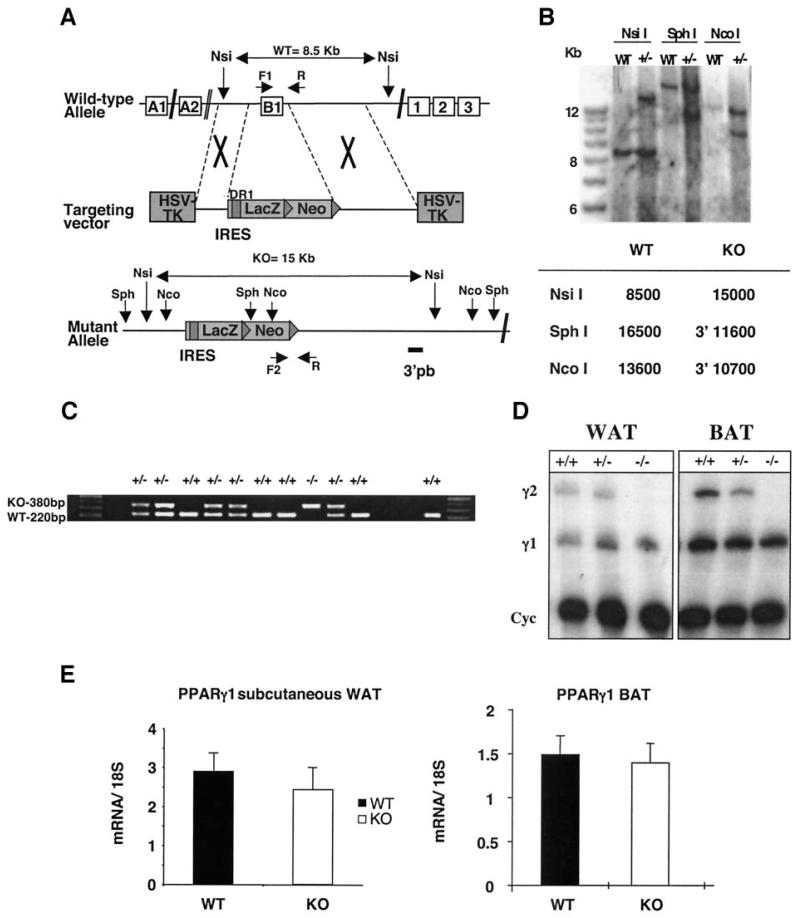
FIG. 1.
Generation of PPARγ2 knockout mouse. A: Schematic representation of PPARγ2 knockout targeting construct and the predicted structure of the recombinant allele. The targeting construct contains 5.2 kb of homologous mouse PPARγ2 genomic DNA, with LacZ/NEO cassette of 5.335 kb. The LacZ/NEO cassette replaces ~164 bp of the PPARγ locus including all 145 bp of exon B1. B: Southern blot analysis of genomic DNA. The 3′ probe (3′pb) is located outside the targeting construct sequence. Genomic DNA was digested with NsiI, SphI, and NcoI. C: Genotyping was performed by multiplex PCR in the same reaction. Specific primers used to detect the knockout (KO) allele were the sense Asc 306 (F2), located in the NEO cassette and PPAR antisense (R) located 42 bp downstream of exon B1 (380-bp PCR product). Primers used to detect the wild-type (WT) allele were the PPAR sense (F1) located 7 bp upstream of exon B1 and PPAR antisense (R) (220-bp PCR product). The binding site for primer F1 was deleted by homologous recombination. D: RPA analysis of PPARγ1 and PPARγ2 in wild-type, heterozygous, and PPARγ2 knockout mice using total RNA from WAT and BAT. Cyc, cyclophilin. E: TaqMan analysis of PPARγ1 in wild-type and PPARγ2 knockout mouse subcutaneous WAT and interscapular BAT.