FIGURE 1.
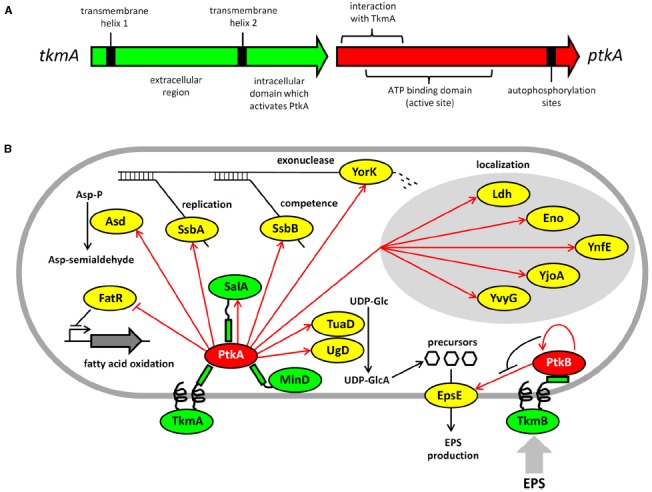
Architecture and protein substrates of B. subtilis BY-kinases. (A) Schematic structure of the ptkA and tkmA genes, encoding the kinase and its activator, respectively. (B) Network of BY-kinases and their substrates in B. subtilis. Proteins are color-coded: BY-kinases are shown in red, kinase activators in green and substrates in yellow. Phosphorylation reactions are indicated with red arrows. Substrate physiological role is indicated next to substrate whose activity is affected by phosphorylation. Substrates whose subcellular localization is affected in the ΔptkA strain are grouped together. The substrate activity is regulated by phosphorylation as follows: Ugd (UDP-glucose dehydrogenase)—activated, Asd (aspartate semialdehyde dehydrogenase)—activated, FatR (repressor of fatty acid oxidation)—inactivated, SsbA and SsbB (ssDNA binding proteins)—activated, YorK (ssDNA exonuclease)—activated, EpsE (glycosyltransferase)—activated.
