Figure 3.
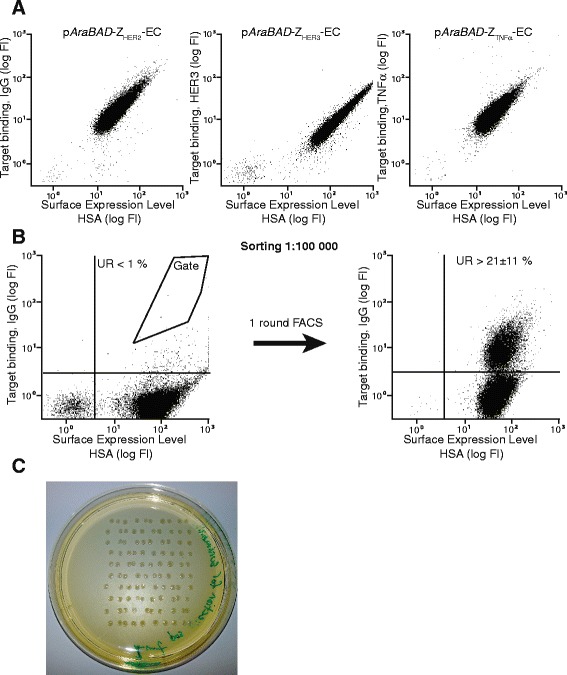
Surface expression of different Affibody molecules on E. coli and fluorescence-activated cell sorting from spiked library. A. Representative dot plots from flow-cytometric analysis of E. coli BL21 (pAraBAD-ZHER2-EC), (pAraBAD-ZHER3-EC) and (pAraBAD-ZTNFα-EC), respectively. Fluorescence intensity corresponding to target-binding on the y-axis and fluorescence intensity corresponding to surface expression level (HSA-binding) on the x-axis. B. Representative dot plots from flow-cytometric sorting of a 1:100,000 mix of E. coli BL21 (pAraBAD-ZIgG-EC) in a background of E. coli BL21 (pAraBAD-ZHER2-EC). Leftmost dot plot shows the mixed population before FACS with the sorting gate indicated in the dot plot. Rightmost dot plot shows the enriched bacteria after sorting and overnight growth. The percentage (mean ± standard deviation) of events in the upper right (UR) quadrant is indicated in the dot plots. The entire experiment was performed in duplicates on different days, using freshly prepared mixtures of bacteria expressing ZHER2 and ZIgG, and freshly prepared reagents. C. Viability test after FACS. Bacterial colonies after sorting 100 (10 x 10 pattern) single bacterial cells directly on a plate containing semi-solid medium with antibiotics and overnight incubation at 37°C. The viability assay was performed in triplicates on different days.
