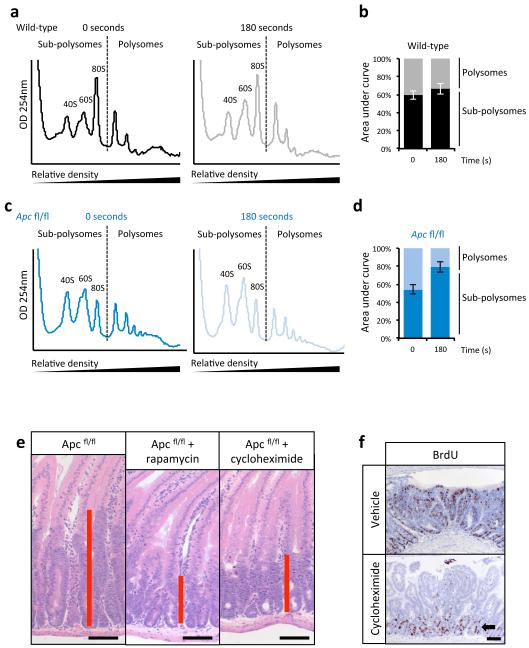
Extended Data Fig 6. Apc deletion increases translational elongation rates and Cycloheximide treatment phenocopies rapamycin treatment.
a) Representative polysome profiles from wild-type ex vivo crypts incubated with harringtonine for 0 seconds (left) and 180 seconds (right) prior to harvest. (n=3 per time point) b) The areas under the sub-polysome (40S, 60S and 80S) and polysome sections as indicated by the dashed lines in a) were quantified and expressed as a percentage of their sum. Data in the bar graph is the average ± s.e.m (n=3 per time point). c) and d) show data for Apc deleted crypts as for wild-type in b) and c) (n=3 biological replicates). e) Representative H+E showing that 35mg/kg cycloheximide treatment phenocopies rapamycin treatment 96hrs after Apc deletion. Treatment began 24hrs after induction (n=3 biological replicates); f) Representative IHC for BrdU showing a loss of proliferation in tumours after 72hrs of 35mg/kg cycloheximide treatment. (n=3 biological replicates). Arrow highlights normal proliferating crypts. Scale bar = 100μm