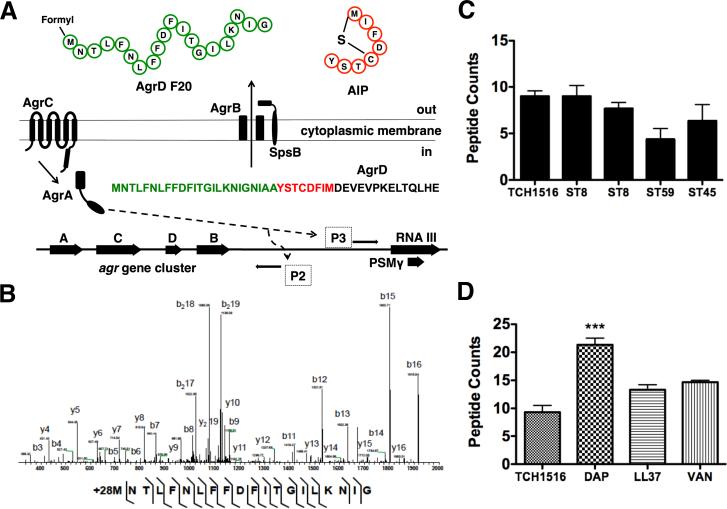
Figure 1.
The accessory gene regulatory system and N-AgrD peptide properties. (a) Four genes, agrABCD comprise the agr biosynthetic network. agrD encodes the peptide substrate undergoes PTM by membrane-bound endopeptidase agrB through cleavage and thiolactone cyclization. Once extracellular, the hybrid molecule consisting of the AIP and the N-terminal region of AgrD is further processed by peptidase SpsB into mature AIP and N-AgrD. Induction of Promoter 2 drives the transcription of the agrABCD operon. Promoter 3 induction results in the expression of RNA III that produces two final products, the primary RNA effector molecule and PSMγ. (b) MS survey of several S. aureus clinical isolates show detectable N-AgrD peptides as peptide counts. (c) Tandem MS sequencing of AgrD F24. Several contingent ion fragments, including reliable mass accuracy, localize a PTM of +28 Da to the initiator methionine of N-AgrD. (d) Antibiotic effects on the number of N-AgrD peptide counts. Challenge with a subinhibitory concentration of daptomycin increased the number of detectable peptides. Statistical analysis performed by one-way ANOVA; ***P < 0.001. Data was expressed as mean ± standard deviation.