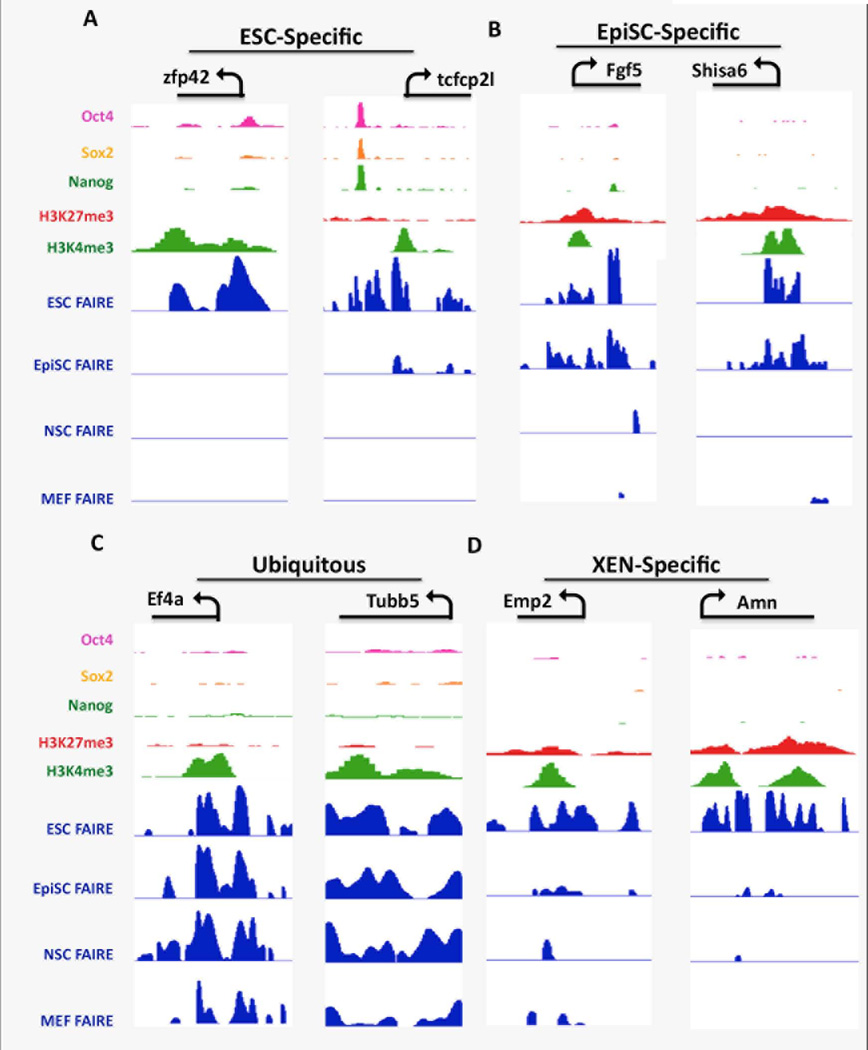
Figure 4. Distinct combinations of FAIRE peaks, Histone modifications, and core TF binding in ESC chromatin at the promoter regions for genes that are ubiquitously expressed, or are differentially expressed in ESCs, EpiSCs, or XEN.
IGV view of genomic regions of representative promoters for (A) genes more highly expressed in ESCs (ESC-Specific), (B) genes more highly expressed in EpiSCs (EpiSC-specific) (C) Ubiquitously expressed genes (Ubiquitous), and (D) genes more highly expressed in XEN (XEN-specific). The transcription unit, TSS and direction of transcription is shown on the top of the panel for each representative gene. FAIRE peaks are shown for each of the cell lines as indicated, along with ChIP-seq data derived from ESCs for H3K4me3- and H3K27me3-modified histones and OCT4, SOX2, and NANOG binding [63, 64]. This and all IGV screen shots shown used the following enrichment score scales: FAIRE tracks 0–10, OSN 0–200, H3K27me3 0–5, H3K4me3 0–30).