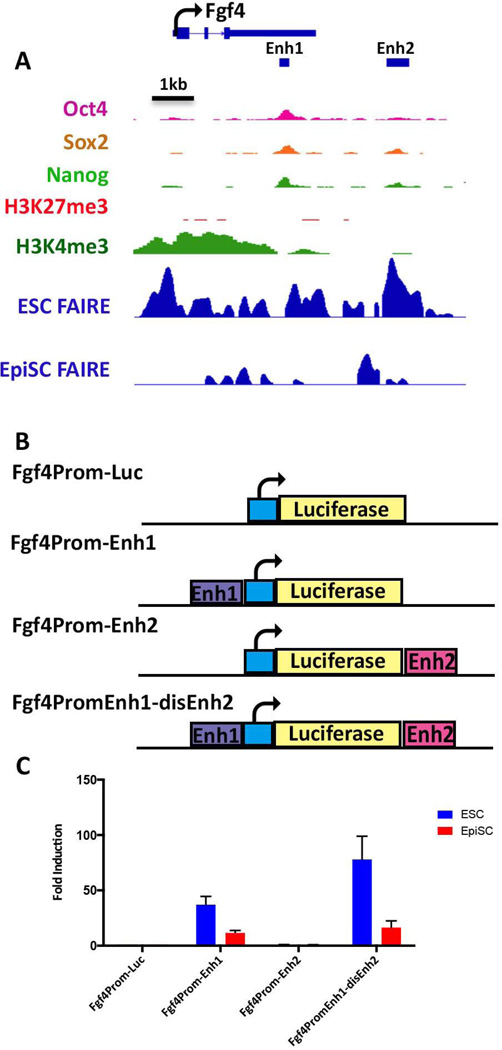
Figure 5. FAIRE-seq reveals an additional ESC enhancer associated with the Fgf4 gene.
(A) IGV view of the Fgf4 locus. The TSS and direction of transcriptional elongation are depicted by the arrow on top of the panel, FAIRE peaks derived from ESCs and EpiSCs are shown along with previously reported ChIP data for the indicated histone modifications and OCT4, SOX2, and NANOG binding in ESCs, [63, 64]. Location of the previously characterized Fgf4 enhancer (Enh1) is shown by the black bar, as well as a unique ESC-specific FAIRE peak, Enh2, located 2 kb downstream of the Fgf4 transcription unit. (B) Schematic depiction of luciferase reporter plasmids. The basal plasmid Fgf4prom-luc [65] consists of a minimal region of the Fgf4 promoter and has little activity on its own. Test plasmids contain either Enh1 or Enh2 inserted a shown within Fgf4prom-Luc, or both enhancers contained within the same plasmid in the case of Fgf4PromEnh1-disEnh2(C) Lysates derived from ESCs or EpiSCs transfected with each of the reporter plasmids indicated on the x-axis were assessed for luciferase activity. Activities are expressed as fold activation relative to that of Fgf4prom-Luc.