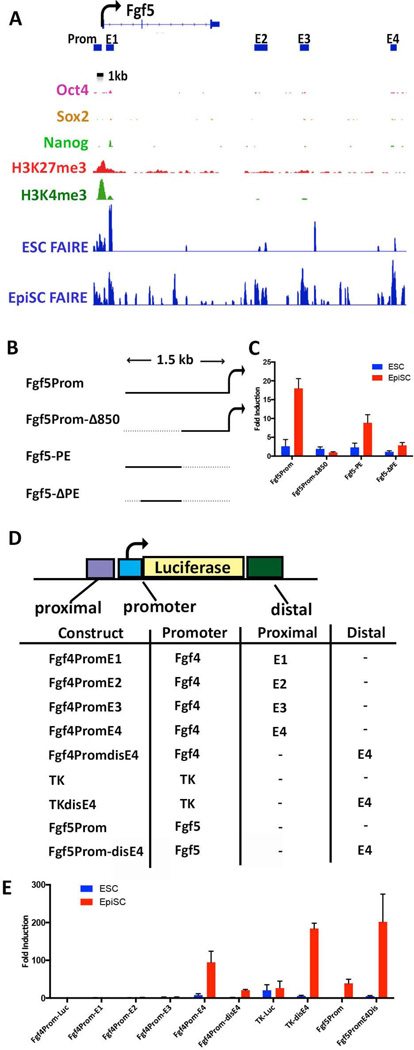
Figure 6. EpiSC-specific enhancers within distal FAIRE sites at the Fgf5 locus.
(A) IGV view of the Fgf5 locus. The TSS and direction of transcriptional elongation are depicted by the arrow on top of the panel. ESC and EpiSC FAIRE peaks are shown along with previously reported ESC ChIP data for the indicated histone modifications and OCT4, SOX2, and NANOG binding [63, 64]. Bars under the Fgf5 gene correspond to the Fgf5 promoter region (Prom) and the locations of distal EpiSC-specific FAIRE peaks (regions E1, E2, E3, E4) that were cloned into luciferase reporter plasmids depicted in (B&D). (B) Fgf5 promoter constructs. Fgf5Prom contains the 1.5 bp DNA region spanning the Fgf5 TSS; the 5’-most 850bp of this fragment are deleted in Fgf5Prom-Δ850. Fgf5-PE contains the 850bp region deleted in Fgf5Prom-Δ850 inserted upstream the minimal promoter in fgf4prom. Fgf5-ΔPE is similar to Fgf5-PE but contains an 18bp deletion of the 5’-most sequences of the 850 DNA fragment. (C) Luciferase activity of the indicated reporter plasmids in ESCs and EpiSCs. Fold activation=relative to that of Fgf4prom-luc. (D) Fgf5 Distal regions. Promoter= the Fgf4 minimal promoter, the Herpes Simplex virus TK promoter [65], or the 1.5kb Fgf5 promoter region as indicated in the table. Fgf4prom-E1-E4 contain regions E1, E2, E3, or E4 upstream of the minimal promoter within Fgf4prom-luc. TKdisE4 and Fgf5Prom-disE4 harbor region E4 distal to the promoters. (E) Luciferase activity of the indicated reporter plasmids in ESCs and EpiSCs. Fold activation=relative to that of Fgf4prom-luc.