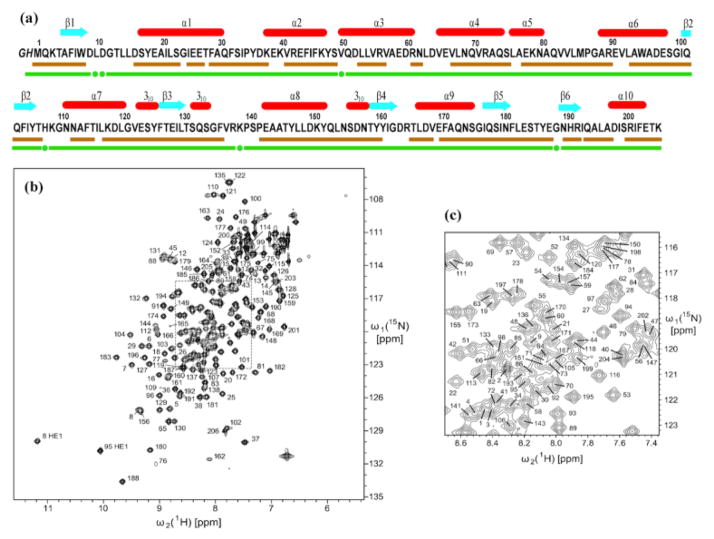
Fig. 1.
Characterization of the solution of the protein NP_346487.1 used for the NMR structure determination and survey of the polypeptide backbone assignments. (a) Amino acid sequence (residues -2 and -1 originate from the expression tag), extent of the automated backbone assignments obtained with the software UNIO-MATCH, which are based on APSY-NMR data alone (brown underline; for some residues only part of the backbone chemical shifts were assigned, as described in the text), and backbone assignments after interactive validation and extension of the automatic assignments with the use of 3D heteronuclear-resolved [1H,1H]-NOESY data (green underline; green dots identify residues with partial assignments of the backbone chemical shifts). Above the sequence, β-strands are indicated by cyan arrows and helices by red bars. (b) 600 MHz 2D [15N,1H]-HSQC spectrum of a 1.5 mM solution of uniformly 13C,15N-labeled NP_346487.1 in 20 mM sodium phosphate buffer at pH 6.5 and T = 298 K. The backbone amide group assignments are indicated by the sequence numbers. (c) Expansion of the central spectral region indicated by broken lines in (b).