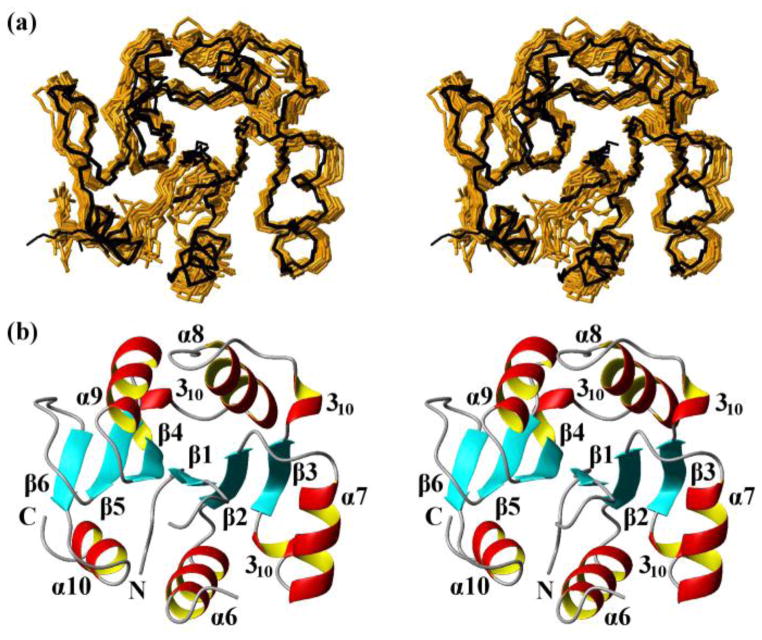
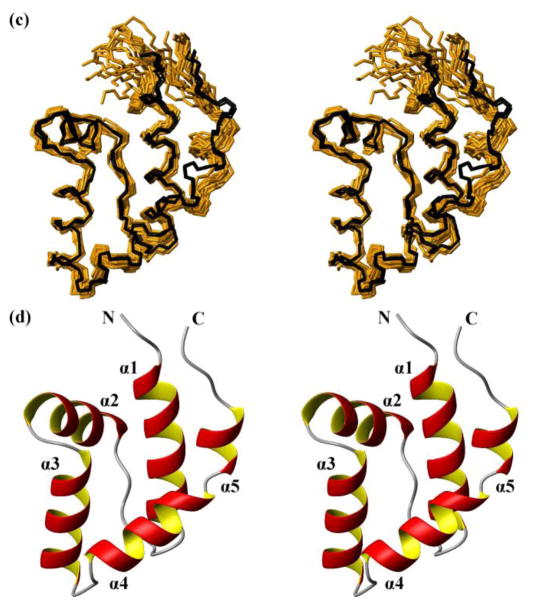
Fig. 3.
NMR structures calculated separately for the two domains of the protein NP_346487.1 from data collected with the intact protein (same data as for Fig. 2, see Table 1; the structures are closely similar to the structures of the individual domains in Fig. 2, see Figure S1 and the text) and comparison with the crystal structure. (a) Core domain (residues 1–10 and 88–206). 20 NMR conformers (brown) and the 4 crystal structures (black) are superimposed for best fit of the polypeptide segments 3–10 and 88–205. (b) Stereo ribbon diagram of the NMR conformer closest to the mean coordinates of the bundle of conformers in (a). Color code: β-strands, cyan; helices, red/yellow; non-regular secondary structure, grey. The individual regular secondary structures are identified, and the two chain ends are marked with N and C. (c) and (d) Cap domain residues 17–80). Same presentations as in (a) and (b), with best-fit superposition of residues 17–80.