Figure 1. Functional predictions of identified proteins.
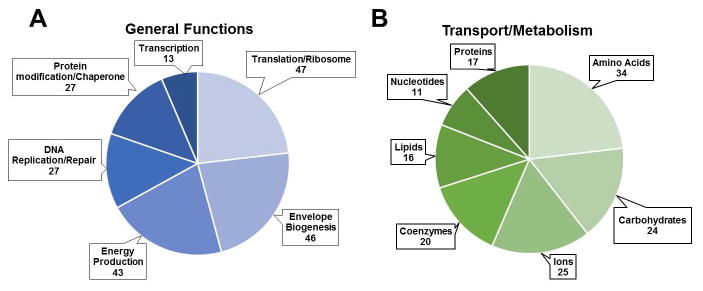
Proteins were assigned to functional groups (COG classes) utilizing the WebMGA server (http://weizhonglab.ucsd.edu/metagenomic-analysis/). Classes containing at least 10 sequences were utilized to generate pie charts of proteins involved in (A) general cellular functions and (B) transport and metabolic functions. COG class names were shortened for clarity. Classes represented are as follows for general cellular functions; Translation/ribosome: J, Envelope biogenesis: M, Energy production: C, DNA replication/repair: L, Protein modification/chaperone: O, Transcription: K, and for metabolism/transport of; Amino acids: E, Carbohydrates: G, Ions: P, Coenzymes: H, Lipids: I, Nucleotides: F, Proteins: U.
