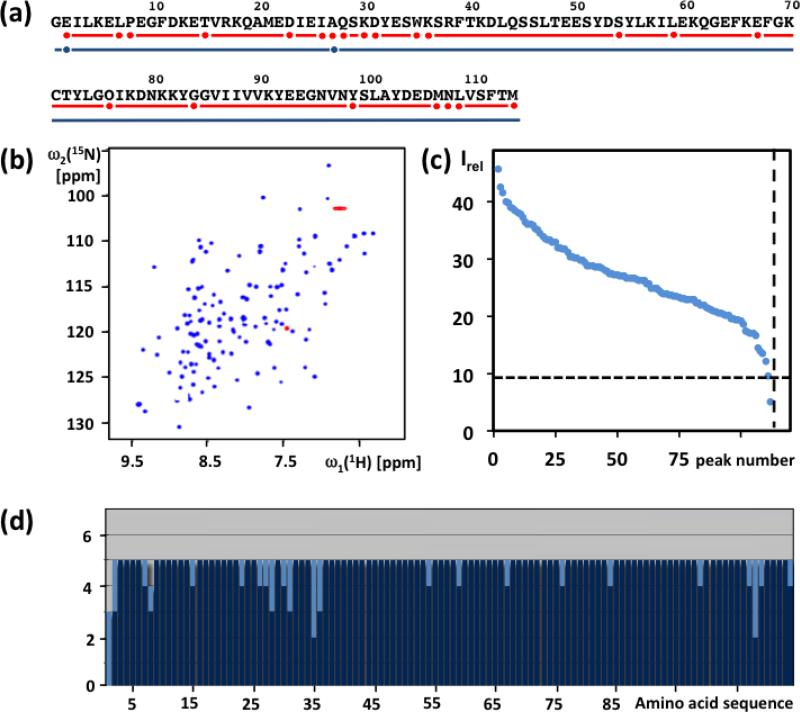
Fig. 1.
Characterization of the solution of the protein ZP_02041089.1 used for APSY-NMR experiments, and result of the automated backbone assignment with UNIO-MATCH-2014. (a) Amino acid sequence (Gly-1 results from the cloning strategy). The red line indicates residues with complete automated backbone assignment by UNIO-MATCH-2014, and red dots identify residues with incomplete assignments. The blue line and the blue dots indicate the corresponding information obtained after interactive validation of the assignments, which are complete except that for E2 and A27 only the chemical shifts of Cα, Hα and Cβ were assigned. (b) [15N,1H]-HSQC spectrum of the 15N- labeled protein recorded at 700 MHz with a 1.7 mm micro-coil probehead. Red color identifies folded peaks of arginine side chain 15N−1H moieties. (c) NMR- profile obtained from the data in (b); the dotted horizontal and vertical lines indicate, respectively, the intensity threshold for detection of APSY-NMR signals (see text) and the number of backbone amide and tryptophan indole 15N−1H correlation signals expected from the amino acid sequence. (d) UNIO-MATCH-2014 output. The dark blue and light blue bars represent the number of assigned and unassigned 1HN, 15N, Cα, Hα and Cβ atoms per residue, respectively.