Figure 4.
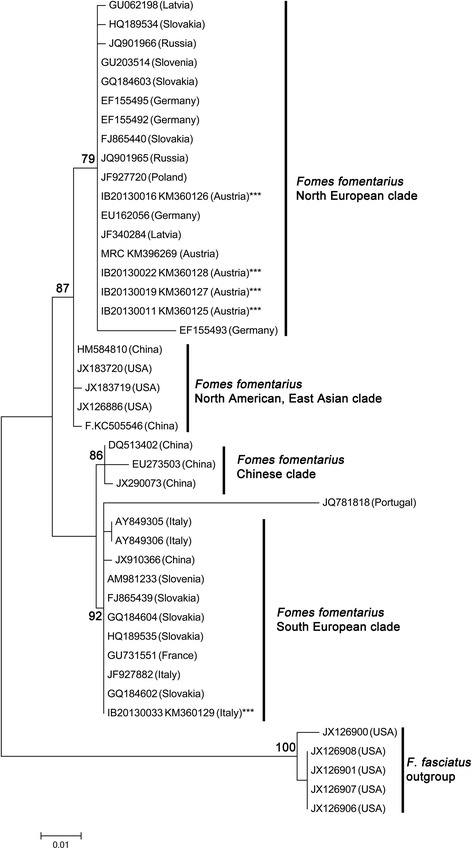
Phylogenetic placement of F. fomentarius strains inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model. The tree with the highest log likelihood (-1266,9966) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 43 nucleotide sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There were a total of 505 positions in the final dataset. Bootstrap values above 75 % are given (1 000 replications). Evolutionary analyses were conducted in MEGA6.
