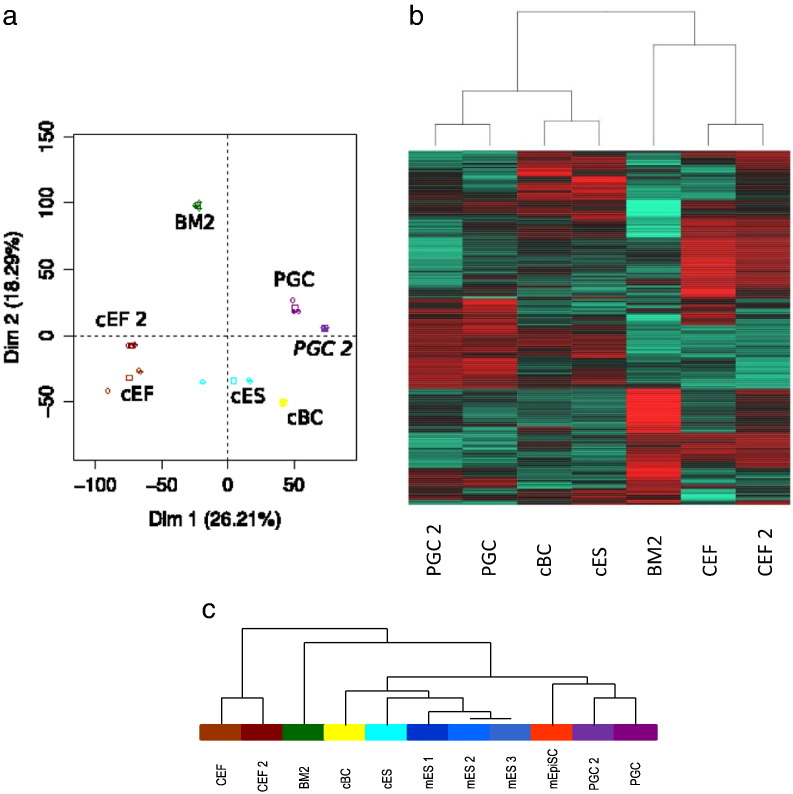
Figure 5.
PCA and heatmap analysis reveals clustering of CEF and PGC with previous data, and cES with cBC (a, b) and with mES in an virtual Array (c).
a: PCA analysis was performed on the 11708 common genes between the data from this study (GSE61221) (PGC, cBC, cES, BM2 and CEF) and those provided by the PGC and CEF samples (PGC 2, CEF 2) of the GSE38168 microarray data as described in Materials and Methods. CEF and PGC from the two studies clustered together. Each sample (circle) is plotted as well as the barycentre of the triplicate (square).
b: The heatmap analysis was performed by ANOVA analysis of the 11708 common genes between the data from this study (GSE61221) and those of the GSE38168 set as listed in A. The heatmap presented was performed with the first 1000 ranked genes.
c:. By incorporating the mouse ES and EpiSC cells in a virtual array approach as described in Materials and Methods, a clustering tree reveals that cES cells cluster more closely with mES cells than with EpiSC, whereas the latter are more closely related to PGC.