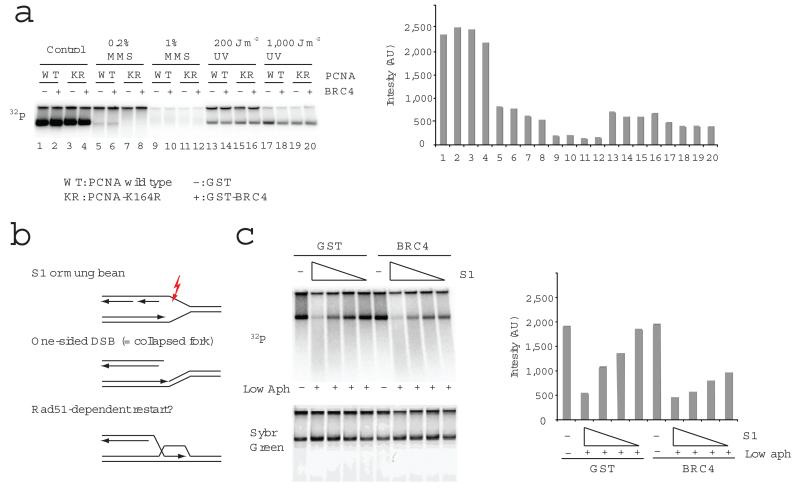
Figure 1.
RAD51 is required for DNA replication in the presence of forks collapsed by a single strand break in the template. (a) The requirement of RAD51 and PCNA modification at Lys 164 for replication of undamaged sperm DNA (control) or MMS or UV treated sperm DNA was tested using GST-BRC4 (BRC4), which sequesters RAD51, and PCNA K164R mutant. Replication products (labelled with 32P-dATP) were resolved by neutral agarose gel electrophoresis and subjected to autoradiography (left). The quantification of the signal is shown in the graph as photon emission intensity (Intensity) expressed in arbitrary units (AU) (right). The data shown here and hereafter represent typical findings of 3 or more experiments. (b) A model for ssDNA specific endonuclease-dependent fork collapse and RAD51-dependent restart. (c) The requirement of RAD51 for DNA replication in the presence of S1 nuclease was tested using sperm nuclei (4000 nuclei per μl) incubated for 80 min in the presence or absence of 1 μg ml−1 aphidicolin (Low aph) and S1 nuclease (0, 2.92, 1.46, 0.73, 0.37 U μl−1). Replication products were detected by autoradiography with quantification shown on the right, as in 1a. Sybr Green staining shows total DNA (Sybr Green).