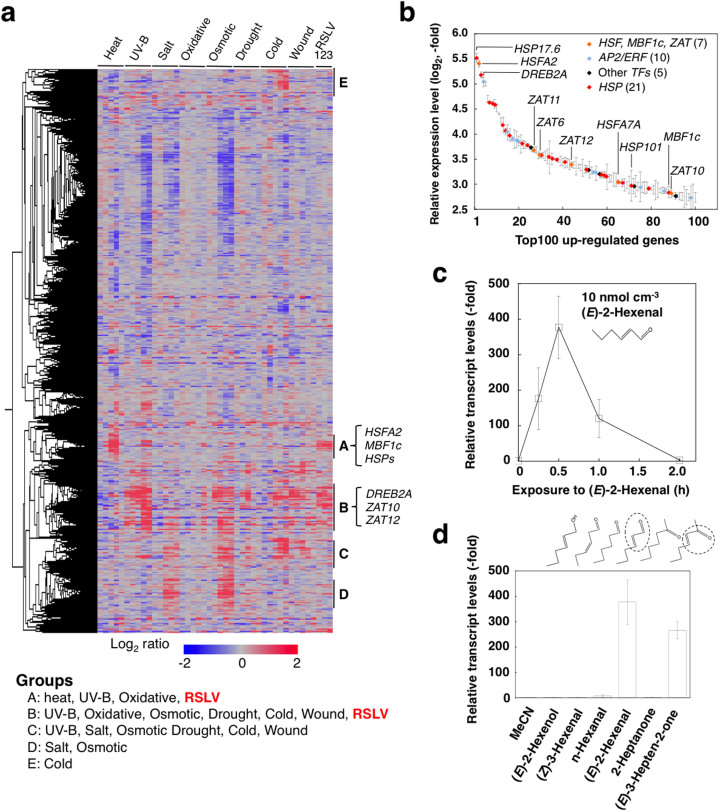
Figure 1. Abiotic stress-related genes were up-regulated by RSLV treatment in Arabidopsis.
(a), Heat map constructed with the whole gene expression. Expression data used are data from shoots collected over the course (0.25, 0.5, 1, 3, 6 h of heat, UV-B, drought or wound stresses) and (0.5, 1, 3, 6, 12 h of oxidative, salt, osmotic or cold stresses) obtained from the AtGenExpress database. RSLVs used for obtaining expression data are (E)-2-hexenal (lane 1), (E)-2-butenal (lane 2) and 3-hepten-2-one (lane 3). Genes induced by heat and various stresses induced were classified into Groups A and B, respectively. In addition, responsive genes to salt, oxidative, osmotic drought, cold and wound (Group C), salt- and osmotic-responsive genes (Group D) and cold-responsive genes (Group E) are shown. (b), Transcription factors and HSPs in the 100 most highly up-regulated genes. Expression was induced by (E)-2-hexenal treatment. Symbols indicate the HSF and ZAT genes (orange), AP2/ERF genes (blue), other transcription factor genes (black), and HSP genes (red). Symbols of other genes are omitted. Data are means ± SE (n = 3). Detailed list is shown in Table S1. (c) and (d), Arabidopsis plants were exposed to (E)-2-hexenal (10 nmol cm−3) for various time periods (0 to 2 h) (c), or series of C6 GLVs and their analogues (each 10 nmol cm−3) for 30 min (d).