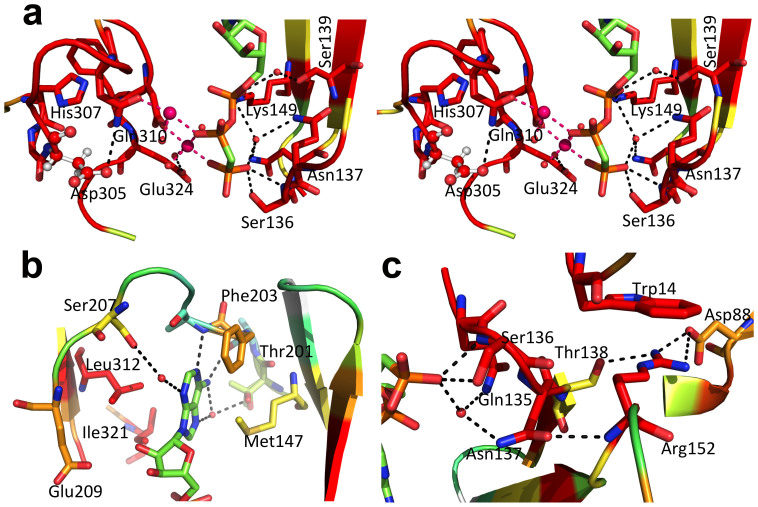
Figure 5. Close view of the AppCp-binding site of MakMvan.
(A) Stereo view of the polar interactions centred on the phosphoryl moieties of AppCp. For all panels, residues are coloured according to conservation as shown in Supplementary Fig. S1, where red corresponds to positions strictly conserved in the actinobacteria maltokinase sequences selected for the multiple alignment. Selected residues are shown as sticks with oxygen atoms in red, nitrogen in blue, sulfur in yellow, phosphorus in orange and carbon in green (nucleotide) or according to sequence conservation (protein). The putative catalytic base (Asp305) is represented as ball-and-stick. Hydrogen bonds are represented as dashed black lines. The magnesium ions are represented as magenta spheres and dashed magenta lines represent bonds to the magnesium coordinating atoms. Red spheres represent ordered water molecules. (B) Detailed view of the interactions with the adenine base of the nucleotide bound to the active site cleft of MakMvan. (C) Close-up of the vicinity of residue Asn137, which bridges the P-loop and the terminal phosphoryl moiety of AppCp to the N-terminal cap domain through the conserved Arg152. Figure prepared with PyMOL (http://www.pymol.org).