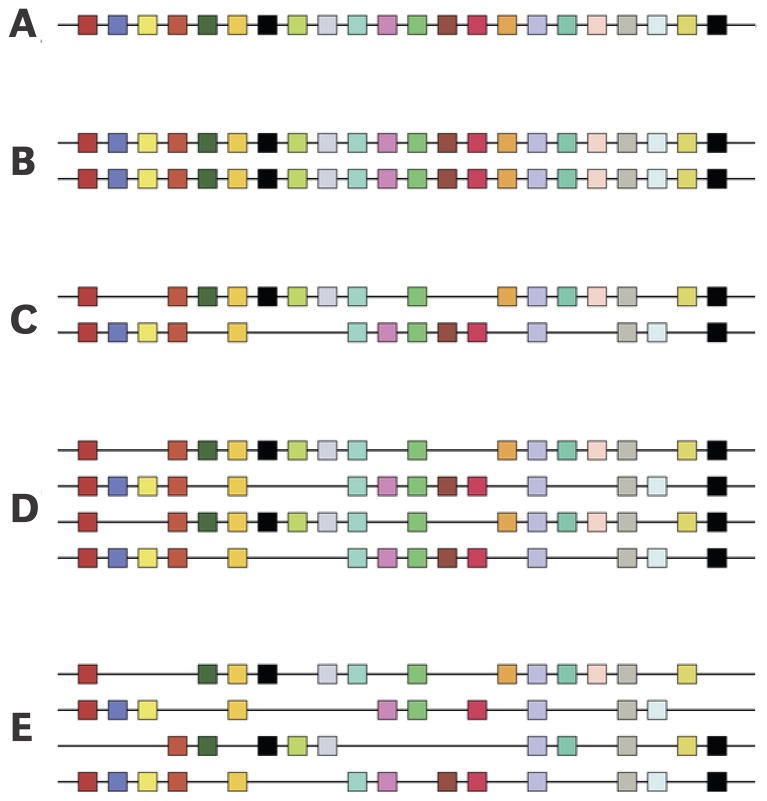
Figure 4.
Diagram depicting the hypothetical effects of two consecutive genome duplications (1R and 2R), as reflected in the physical linkage arrangement of paralogous genes – specifically, the four-fold pattern of intragenomic synteny. (A) Hypothetical chromosome in the vertebrate common ancestor; (B) The first genome duplication produces a complete set of paralogs in identical order; (C) Many paralogous gene copies are subsequently deleted from the genome; (D) The second genome duplication produces yet another set of paralogs in identical order, with multigene families that retained two copies now present in four. (E) With the passage of time, additional gene losses ensue, thereby obscuring the four-fold pattern of synteny. Modified from Dehal and Boore (2005).