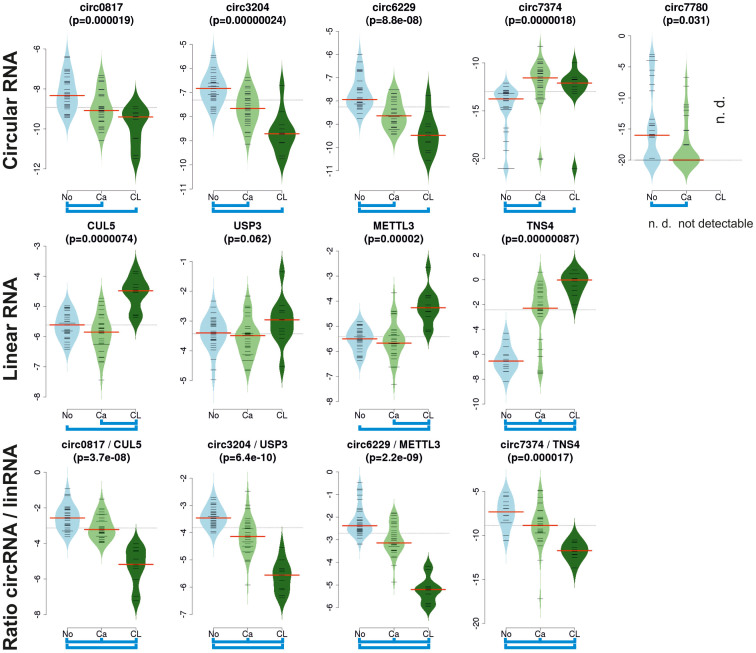
Figure 3. Ratio of circular to corresponding linear RNAs in 31 matched normal mucosa (No) and colorectal tumour (Ca) tissues and in eleven colorectal cancer cell lines (CL).
Expression of (i) five exemplarily chosen circular RNAs (circ0817, circ3204, circ6229, circ7374, and circ7780), (ii) four corresponding linear RNAs (CUL5, USP3, METTL3, and TNS4), and (iii) ratios of circular to linear RNAs (circRNA/linRNA); determined by RT-qPCR. Circ7780 was not detectable (n.d.) in cancer cell lines. Y-axes represent relative expression values normalised to housekeeping genes for the upper two panels and the ratios of circular RNAs to linear RNAs in the lower panel. Significance for multiple comparison of No versus Ca versus CL was assessed by Kruskal-Wallis rank sum tests (given p-values) followed by pairwise comparisons applying the post-hoc Nemenyi test with Chi-squared approximation (significant pairs indicated by blue brackets).