Figure 1. Proportion of correct reads for the three genetic systems (simple: a single allele per individual, squares; medium: two alleles, circles; and complex: multiple alleles, triangles) using standard PCR conditions (open) and modified PCR conditions to reduce chimera formation (gray).
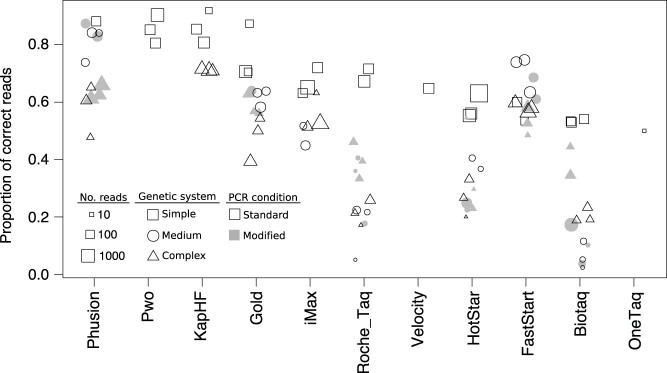
The size of the shape is indicative of the number of reads (see legend). All enzymes yielded at least 50% correct reads in the simplest system, mitochondrial DNA (Test 1; open squares). Some enzymes only worked for a given set of conditions (cycling conditions/genetic system). A group of enzymes consisting of Phusion, Gold and FastStart yielded a high proportion of correct reads cosistently accross all conditions. Others, such as Roche Taq, HotStar and Biotaq, yielded a low percent of correct reads for the more complex systems (MHC class I and MHC class II). Abbreviations as defined in Table 1.
