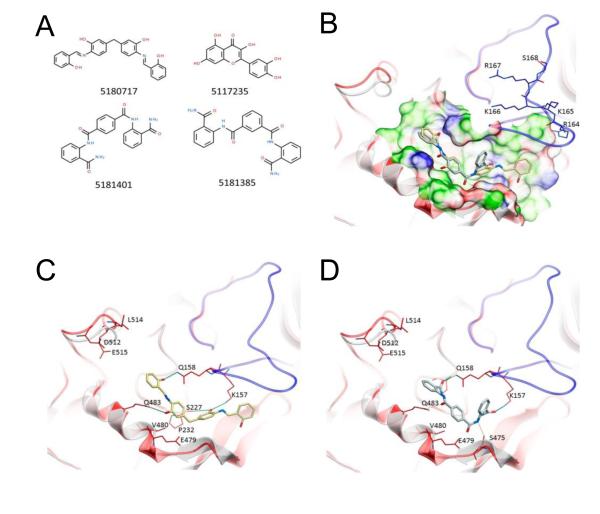
Figure 2.
Structures of small-molecule anthrax toxin inhibitors and their modes of binding to PA. (A) The compounds investigated in ref (72). (B) PA crystal structure 1T6B (red ribbon) superimposed on the crystal structure 3TEW (gray ribbon) with the ordered furin loop in 3TEW highlighted in blue. The furin-type protease cleaves after the sequence 164RKKR, which is shown in stick representation with carbon atoms colored blue. The predicted binding poses for inhibitors 5180717 and 5181401 are displayed in stick representation with carbon atoms colored yellow and gray, respectively. The binding pocket surface for 1T6B used for virtual screening is displayed (white = neutral surface, green = hydrophobic surface, red = hydrogen bonding acceptor potential, blue = hydrogen bond donor potential). (C, D) Predicted interactions of 5180717 (yellow stick) and 5181401 (gray stick), respectively, with PA. Both ligands make common hydrogen bonds with Q158, Q483, and K157. Reprinted with permission from ref (72). Copyright 2012. American Chemical Society.