Figure 1. The RNA-directed DNA methylation pathway (RdDM) in Arabidopsis.
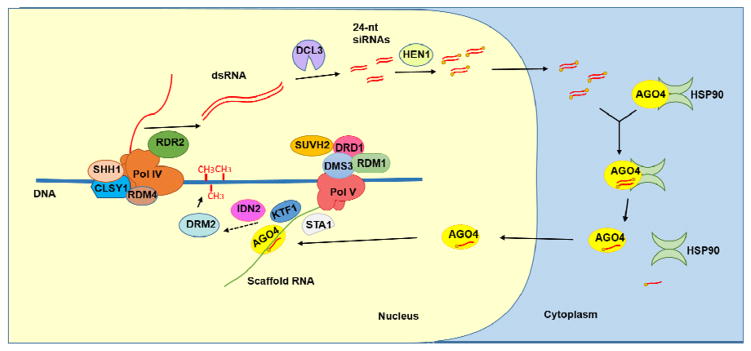
At the early stage of RdDM, RNA Polymerase IV (Pol IV) is proposed to generate a single-stranded non-coding RNA, which is converted into double-stranded RNA (dsRNA) by RNA-DEPENDENT RNA POLYMARASE 2 (RDR2). SAWADEE HOMEODOMAIN HOMOLOG 1 (SHH1), CLASSY1 (CLSY1), RNA-DIRECTED DNA METHYLATION 4 (RDM4) and RDR2 were found to be in the same complex as Pol IV and may assist the recruitment of, or transcription by, Pol IV. 24-nt siRNA duplexes are produced from dsRNA precursors by DICER-LIKE 3 (DCL3) and are methylated at their 3′ ends by HEN1. The 24-nt siRNAs are exported into the cytoplasm to be loaded into AGONAUTE4 (AGO4). The molecular chaperon HSP90 facilitates the release of the siRNA passenger strand. The AGO4 protein loaded with an siRNA is translocated into the nucleus. RNA POLYMERASE V (Pol V) generates a non-coding scaffold RNA at RdDM loci. SU(VAR)3-9 HOMOLOG 2 (SUVH2) or its homolog SUVH9 contributes to the recruitment of Pol V to methylated DNA. A protein complex containing at least three proteins, DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1), DEFECTIVE IN MERISTEM SILENCING 3 (DMS3) and RNA-DIRECTED DNA METHYALTION 1 (RDM1), is also required to recruit Pol V to target loci. A splicing factor STABILIZED 1 (STA1) is required for the production of scaffold RNAs, although its mechanism of action is unclear. The scaffold RNA is able to recruit the silencing effector AGO4 as well as KOW DOMAIN-CONTAINING TRANSCRIPTION FACTOR 1 (KTF1), which may help to recruit downstream factors of RdDM. A dsRNA binding protein INVOLVED IN DE NOVO 2 (IDN2) is proposed to recognize the complex of the siRNA and the scaffold RNA. These events further recruit DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) to the RdDM loci through an unknown mechanism.
