SUMMARY
RUNX3 functions as a tumor suppressor in the gastric epithelium, where its inactivation is frequently observed during carcinogenesis. We identified IL23A as a RUNX3 target gene in gastric epithelial cells. This was confirmed in a series of in vitro analyses in gastric epithelial cell lines. In elucidating the underlying regulatory network, we uncovered a prominent role for the TNF-α/NF-κB pathway in activating IL23A transcription. Moreover, the activating effect of TNF-α was markedly augmented by the infection of Helicobacter pylori, the primary cause of human gastritis. Of note, H. pylori utilized the CagA/SHP2 pathway to activate IL23A, as well as the induction of the NOD1 pathway by iE-DAP. Importantly, RUNX3 synergized strongly with these physiologically relevant stimuli to induce IL23A. Lastly, we present evidence for the secretion of IL23A by gastric epithelial cells in a form that is distinct from canonical IL-23 (IL23A/IL12B).
INTRODUCTION
In the stomach, persistent infection by the gastric pathogen Helicobacter pylori and the ensuing inflammation is the single greatest risk factor for gastric neoplasia (Fox and Wang, 2007). While much is known of the mechanisms and consequences of H. pylori infection, little is known of the innate immunity within gastric epithelial cells that acts as the host’s foremost defense (Monack et al., 2004).
Innate immunity in the mucosa is founded on a tissue niche of epithelial, stromal, and hematopoietic cells, where cell-to-cell communication is dependent on a complex network of immune signaling. Of prime importance is the NF-κB pathway, which plays a cardinal role in mediating tissue inflammation in response to pathogen infection, physical insults, and proinflammatory cytokines, such as tumor necrosis factor α (TNF-α) and interleukin-1 (IL-1) (Jobin and Sartor, 2000). A key epithelial response to H. pylori infection is the secretion of the chemokine IL-8, which recruits leukocytes for the prompt clearance of pathogens (Censini et al., 1996). While IL-8 is an important component of host response against infection, the full range of immune signals released by infected gastric epithelial cells remains to be determined.
As the causative relationship between inflammation and cancer becomes increasingly established, evidence has emerged that classical tumor suppressors can influence inflammation and immunity through crosstalk, such as those between the p53 and NF-κB pathways (Baldwin, 2012). The Runt-related transcription factor RUNX3 is a well-established tumor suppressor in the gastric epithelium, where its inactivation is observed in up to 80% of primary gastric tumors (Ito et al., 2005; Li et al., 2002). In mice, genetic ablation of Runx3 leads to the development of spasmolytic polypeptide expressing metaplasia (SPEM), a pre-neoplastic condition often associated with H. pylori infection in humans (Ito et al., 2011). In addition to these epithelial cell-autonomous functions, Runx3 is a key player in hematopoiesis and, together with Runx1, is essential for the proper differentiation and functioning of T cells, B cells, natural killer cells, and myeloid lineages (Collins et al., 2009; Levanon et al., 2014; Puig-Kröger and Corbí, 2006; Watanabe et al., 2010).
In this study, we describe a role for RUNX3 in the direct regulation of IL23A in strong cooperation with TNF-α/NF-κB and H. pylori infection in gastric epithelial cells. Our data further suggest the secretion of IL23A in a form that appears distinct from canonical IL23A/IL12B. Consistent with these findings, we detect the expression of IL23A, independent of IL12B, in purified human primary gastric epithelial cells. These findings implicate IL23A to be a component of the epithelial innate immune response, whose expression is strongly dependent on the tumor suppressor RUNX3.
RESULTS
IL23A Is Transcriptionally Regulated by RUNX3 in Gastric Epithelial Cells
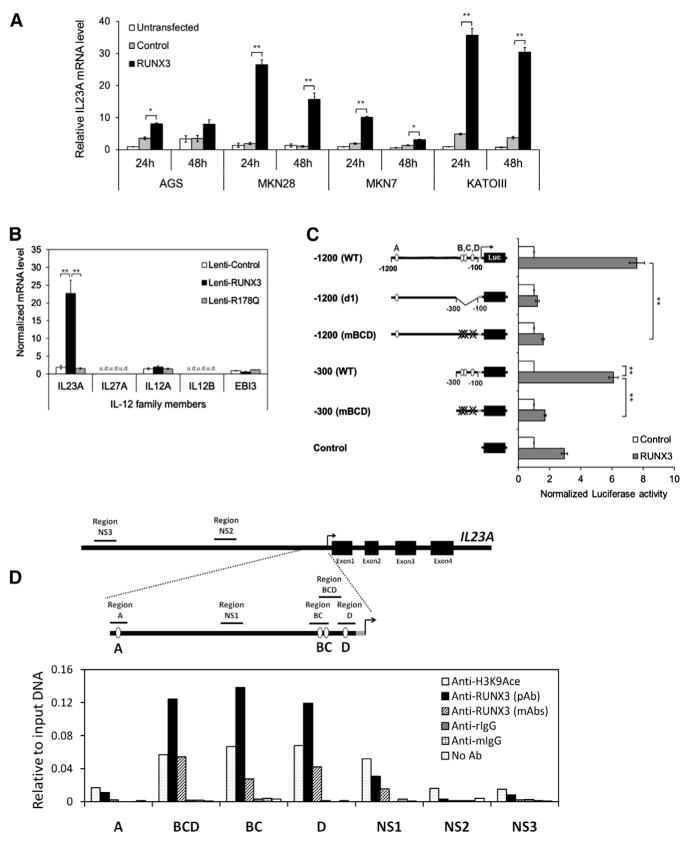
In an expression microarray study, IL23A was identified as a putative target gene of RUNX3 in AGS gastric carcinoma cells (J.K.W.K., D.C.-C.V., and Y.I., unpublished data). This was confirmed in a number of RUNX3-negative human gastric carcinoma lines, demonstrating an important role for RUNX3 (Figure 1A). To investigate if RUNX3 acts transcriptionally on IL23A and whether it has similar effects on other IL-12 family members, AGS cells were transduced with lentivir-uses expressing wild-type RUNX3 or DNA-binding-defective RUNX3R178Q (hereafter Lenti-RUNX3 and Lenti-RUNX3R178Q) and analyzed by quantitative RT-PCR (qRT-PCR). This revealed that RUNX3 specifically induced the expression of IL23A in a DNA-binding-dependent manner while having no effect on other IL-12 family members (Figure 1B). Of note, the expression of IL12A, IL27A, and IL12B was very low or undetectable in this cell type (Figure 1B). To study the molecular mechanism underlying the induction of IL23A, a promoter fragment spanning −1,200 to +105 of the IL23A locus (Figure S1A) was cloned into a firefly reporter construct (hereafter IL23A-1200 reporter). Transient transfection of IL23A-1200 reporter, together with an expression vector encoding RUNX3, into KATOIII and other gastric lines resulted in an induction in luciferase activity, indicating that the cloned promoter fragment recapitulates the transactivating effect of RUNX3 (Figure 1C). By a combination of sequence analysis and empirical mapping, it was determined that three proximal RUNX sites, two of which are noncanonical, are necessary for RUNX3’s transactivation of the IL23A promoter (Figure 1C; Figures S1B and S1C). Notably, the non-canonical site D appeared particularly important for the full effects of RUNX3, while the distal site A appeared nonfunctional (Figure S1C).
Figure 1. IL23A Is Transcriptionally Regulated by RUNX3 in Gastric Epithelial Cells.
(A) IL23A mRNA expression was induced by exogenous RUNX3 in multiple RUNX3-negative gastric cancer cell lines. GFP-positive transfected cells were enriched by FACS at 24 hr and 48 hr posttransfection and analyzed by qRT-PCR. Normalized IL23A levels are expressed relative to untransfected control values.
(B) RUNX3 specifically induced IL23A in gastric epithelial cells. AGS cells transduced with the indicated viruses were analyzed by qRT-PCR for the expression of the IL-12 family of cytokine genes. Normalized data are presented relative to the Lenti-control sample (mean ± SEM; n = 3) (u.d., undetected).
(C) RUNX3 mediates its effect through the proximal RUNX sites B, C, and D of the IL23A promoter. Mutation and deletion variants of the IL23A-1200 reporter construct were transiently transfected into KATOIII cells together with either control or RUNX3 expression vectors. Normalized luciferase activities are expressed relative to the values of control samples for each construct. Data presented are derived from independent biological triplicates (mean ± SEM).
(D) Physical occupancy of RUNX3 on the IL23A promoter in Lenti-RUNX3-transduced AGS cells was detected by ChIP analysis using polyclonal or monoclonal RUNX3-specific antibodies. Enrichment of the indicated genomic fragments was detected by qPCR (Table S1) and expressed relative to input DNA. Nonspecific immunoglobulin G from rabbit (rIgG) and mouse (mIgG) served as negative controls; H3K9Ace antibody was used as a positive control. NS, nonspecific region; pAb, polyclonal antibody; mAbs, monoclonal antibodies. *p < 0.05; **p < 0.01.
See also Figure S1.
To demonstrate the in vivo occupancy of these functional sites by RUNX3, chromatin immunoprecipitation (ChIP) assays were performed on AGS cells that had been transduced with Lenti-RUNX3 (Figure 1D). Polyclonal and monoclonal RUNX3 antibodies strongly enriched genomic DNA fragments bearing sites B, C, and D. Consistent with reporter assay data, genomic fragments bearing site A were not significantly enriched. Similarly, no enrichment by RUNX3-specific antibodies was observed for regions that contain no RUNX sites (Figure 1D). Comparable results were obtained in HFE-145, an untransformed gastric epithelial line, as well as in THP-1, a myeloid cell line in which IL23A could be strongly induced by lipopolysaccharide (LPS) (Figures S1D–S1F). Taken together, these cellular and biochemical data provide strong evidence that RUNX3 transactivates IL23A promoter in gastric epithelial cells through its direct binding to sites B, C, and D.
RUNX3 Augments TNF-α Induction of IL23A Expression
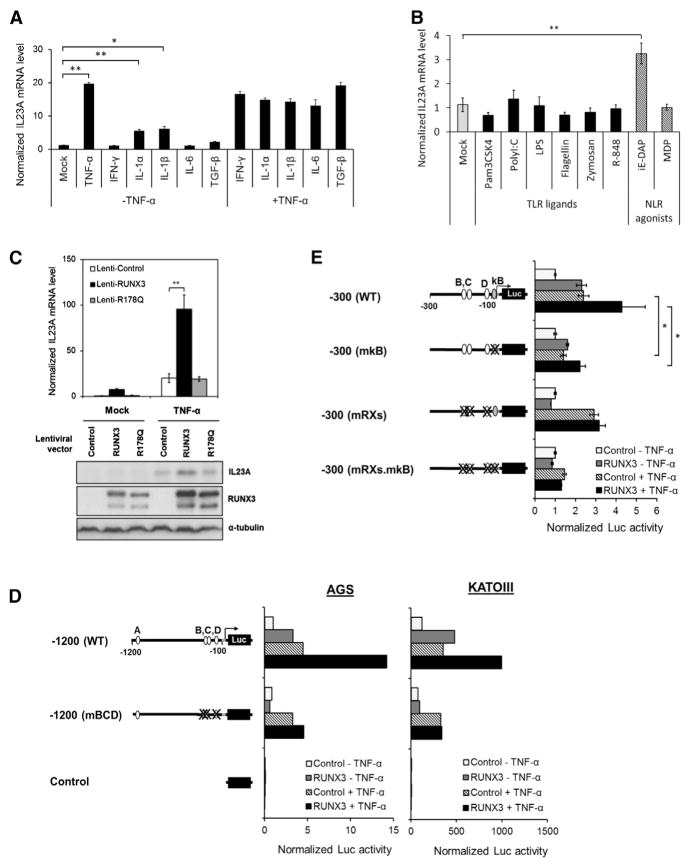
To evaluate the participation of NF-κB, STAT3, and SMAD3, which are known to regulate IL23A in myeloid cells, AGS gastric epithelial cells were treated with stimuli upstream of these transcription factors. This resulted in the clear induction of IL23A by TNF-α, IL-1α, and IL-1β, indicating the involvement of NF-κB (Figure 2A). In contrast, interferon-γ (IFN-γ), IL-6, and TGF-β had no effect on IL23A expression (Figure 2A). A role for NF-κB is further supported by the significant induction of IL23A mRNA by the NOD1 agonist D-glutamylmeso-diaminopimelic acid (iE-DAP) (Figure 2B). Interestingly, IL23A expression in AGS and SNU16 cells was refractory to LPS, a potent inducer of IL23A in THP-1 cells, and all other TLR/NLR agonists tested (Figure 2B; Figures S1F and S2).
Figure 2. RUNX3 Augments TNF-α Induction of IL23A Expression.
(A and B) The effects of various proinflammatory stimuli on the expression of IL23A. AGS cells were treated with the indicated (A) cytokines and (B) ligands and agonists of various TLRs and NLRs for 6 hr prior to qRT-PCR measurement of IL23A transcript. Normalized IL23A mRNA levels are expressed relative to that of untreated controls (Mock). Data presented are derived from biological triplicates (mean ± SEM).
(C) Cooperative induction of IL23A by RUNX3 and TNF-α. AGS cells were transduced with the indicated lentiviruses for 48 hr and treated with TNF-α for 6 hr for qRT-PCR analysis (top) or 18 hr for western blot analysis (bottom). Top: normalized IL23A mRNA levels are plotted relative to the basal values of Lenti-control-infected cells (mean ± SEM; n = 3). Bottom: whole-cell lysates were analyzed for IL23A, RUNX3, and α-tubulin (loading control) expression by western blotting.
(D and E) Sequence requirement for TNFα induction of IL23A promoter. Wild-type and mutant variants of the IL23A promoter reporter constructs were transiently transfected into the cells together with either control or RUNX3 expression vector for 24 hr followed by TNF-α treatment for 24 hr. Normalized luciferase activities are presented relative to those of untreated control samples (mean ± SEM; n = 3). *p < 0.05; **p < 0.01.
See also Figure S2.
To further understand the relationship between RUNX3 and TNF-α, AGS cells were transduced with Lenti-RUNX3 and Lenti-RUNX3R178Q and treated with TNF-α. The ectopic expression of RUNX3, but not RUNX3R178Q, markedly augmented TNF-α induction of IL23A mRNA and protein (Figure 2C). To demonstrate that the partnership between TNF-α and RUNX3 occurs at the level of transcriptional activation, reporter assays were performed. This revealed that the activating effects of TNF-α on the IL23A-1200 reporter were enhanced by exogenous RUNX3 in a manner dependent on the proximal RUNX sites (Figure 2D). In a reciprocal experiment, mutation of a functional NF-κB site abolished TNF-α responsiveness and cooperation between RUNX3 and TNF-α (Figure 2E). Furthermore, the mutant construct IL23A-300mκB was less responsive to RUNX3, suggesting that RUNX3’s action is partly dependent on binding of NF-κB (Figure 2E). Together, these data demonstrate a functional cooperation between RUNX3 and NF-κB in the transactivation of IL23A promoter in gastric epithelial cells.
H. pylori Is a Potent Inducer of IL23A in Gastric Epithelial Cells
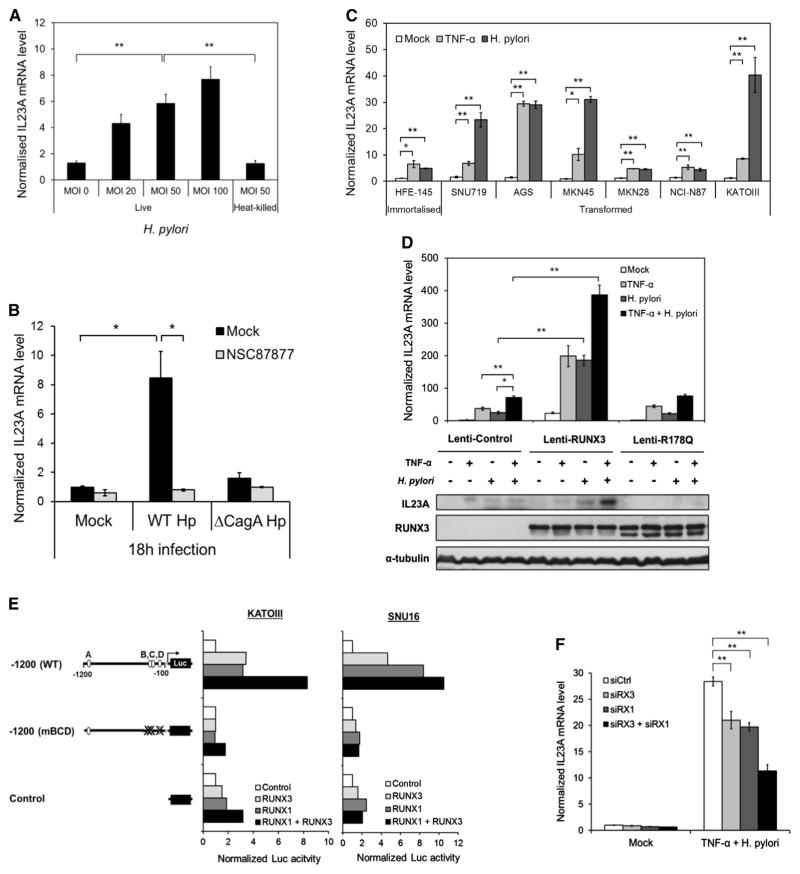
Intracellular delivery of iE-DAP is a consequence of H. pylori infection. To ascertain the physiological significance of the induction of IL23A by iE-DAP (Figure 2B), AGS cells were infected with H. pylori at increasing multiplicity of infection (MOI). This resulted in a dose-dependent induction of IL23A (Figure 3A). Notably, heat-killed H. pylori had no effect, which is consistent with the reliance of an active type IV secretion system (T4SS) for the intracellular transportation of iE-DAP (Figure 3A). As the H. pylori oncoprotein CagA is also delivered by the T4SS and known to activate host gene expression, its effect on IL23A was also investigated. A comparison in the induction kinetics of wild-type CagA-positive H. pylori (WT Hp) and its isogenic CagA-deficient mutant (ΔCagA Hp) revealed that sustained induction of IL23A in infected cells requires CagA (Figure S3A). This requirement was confirmed by the strong induction of IL23A by CagA-proficient H. pylori, but not its ΔCagA counterpart, at the later time point of 18 hr (Figure 3B). Importantly, this induction was completely blocked by an inhibitor of SHP-2 phosphatase (NSC87877), concordant with CagA’s known ability to utilize the SHP-2/ERK pathway (Figure 3B).
Figure 3. H. pylori Is a Potent Inducer of IL23A in Gastric Epithelial Cells.
(A) H. pylori activates IL23A in a dose-dependent manner. AGS cells were cocultured with different MOIs of live or heat-killed H. pylori for 6 hr prior to qRT-PCR quantification of IL23A transcript. The normalized values are presented relative to those of uninfected control samples (MOI 0) (mean ± SEM; n = 3).
(B) IL23A induction by H. pylori requires activation of the SHP-2/ERK pathway by oncoprotein CagA. AGS cells were preincubated with 50 μM of SHP-2 inhibitor (NSC87877) for 3 hr followed by infection with wild-type or ΔCagA strains of H. pylori at MOI100 or vehicle (Mock) for 18 hr. The normalized values are presented relative to those of uninfected control samples (mean ± SEM; n = 3).
(C) TNF-α and H. pylori activate IL23A in a diverse range of gastric epithelial cell lines. Transformed and untransformed gastric epithelial cell lines were stimulated with TNF-α (10 ng/ml) or wild-type H. pylori (MOI100) for 6 hr. IL23A mRNA levels were determined by qRT-PCR and normalized values are presented relative to values of untreated controls of each cell line (mean ± SEM; n = 3).
(D) The expression of IL23A in response to RUNX3, TNF-α, and H. pylori. AGS cells were transduced with the indicated lentiviruses for 48 hr prior to treatment with TNF-α and/or H. pylori for 6 hr for qRT-PCR analysis (top) or 18 hr for western blot analysis (bottom). Top: normalized IL23A levels are presented relative to those of untreated AGS cells infected with Lenti-control virus (mean ± SEM; n = 3). Bottom: whole-cell lysates were analyzed for IL23A, RUNX3 and α-tubulin protein expression by western blotting.
(E) Both RUNX1 and RUNX3 transactivate the IL23A promoter in gastric epithelial cells. KATOIII and SNU16 were transiently transfected with the IL23A-1200 reporter construct together with the indicated expression vectors for 48 hr. Normalized reporter activities of each construct are plotted relative to the values of corresponding control samples.
(F) The effects of RNAi knockdown of RUNX3 and RUNX1 on IL23A expression. HFE-145 cells were transfected with control small interfering RNA (siRNA) (siCtrl), RUNX3 siRNA (siRX3), and/or RUNX1 siRNA (siRX1) for 48 hr prior to treatment with H. pylori and TNF-α for 6 hr and qRT-PCR measurements. Normalized IL23A levels are plotted relative to those of untreated siCtrl sample (mean ± SEM; n = 3). *p < 0.05; **p < 0.01.
See also Figure S3.
To determine if the effects of H. pylori and TNF-α are conserved across gastric epithelial cells, a range of immortalized or transformed gastric epithelial cell lines of various origins was subjected to treatment. This revealed that IL23A expression was significantly induced by TNF-α and H. pylori in all cell lines tested, indicating that the molecular mechanisms underlying their activities are well conserved and intrinsic to gastric epithelial cells (Figure 3C).
As TNF-α and H. pylori each play important roles in the etiology of inflammation-associated gastric cancer, a potential cooperation between these distinct signals and RUNX3 was investigated. Lentivirus-transduced AGS cells were subjected to individual or combined treatments of TNF-α and H. pylori. As in the case of TNF-α, expression of exogenous RUNX3, but not RUNX3R178Q, greatly enhanced the induction of IL23A in response to H. pylori infection (Figure 3D, top panel). Importantly, cotreatment with TNF-α and H. pylori resulted in stronger induction, which was further enhanced by wild-type RUNX3. The increases in IL23A expression were confirmed by western blotting, revealing RUNX3 as a critical determinant in cellular response to TNF-α and H. pylori (Figure 3D, bottom panel). Collectively, these data indicate that IL23A is part of the host response in gastric epithelial cells in reaction to inflammation and infection. It also implicates a role for RUNX3 in orchestrating this response. Of note, RUNX3’s action appears specific to a distinct subset of genes, as it had no effect on H. pylori induction of IL8, a well-established H. pylori target gene (Figure S3B).
As RUNX1 is also expressed in gastric epithelial cells, its ability to transactivate IL23A promoter was tested. In KATOIII and SNU16 cells, the cotransfection of a RUNX1 expression vector resulted in the RUNX sites-dependent induction of IL23A-1200 reporter activities comparable to that of RUNX3 (Figure 3E). To confirm the participation of both RUNX proteins, RNAi knockdown of RUNX1 or RUNX3 was performed in HFE-145 cells, which resulted in weaker induction of IL23A by H. pylori and TNF-α (Figure 3F; Figures S3C and S3D). Moreover, concurrent targeting of both RUNX proteins had additive effects, indicating overlapping and nonredundant functions (Figure 3F). These data indicate a similarly important role for RUNX1 in the transcriptional regulation of IL23A in response to TNF-α/H. pylori.
Epithelial IL23A Is Secreted in a Form Distinct from Heterodimeric IL-23
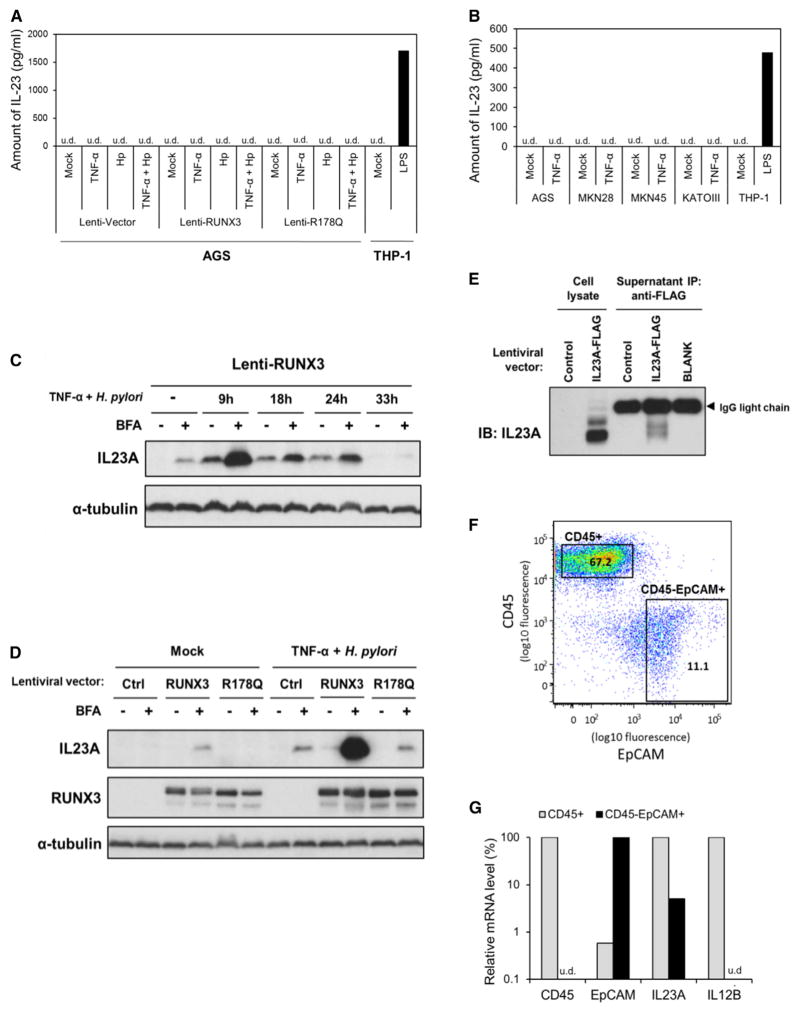
IL23A interacts with IL12B to form the heterodimeric IL-23. However, despite robust expression of IL23A, IL12B is essentially undetectable by qRT-PCR in all gastric carcinoma lines tested (data not shown). The absence of IL12B is confirmed by the inability of two different commercially available ELISA kits to detect heterodimeric IL-23 (IL23A/IL12B) in the supernatants of maximally induced AGS cells (Figure 4A) and several TNF-α activated gastric carcinoma lines (Figure 4B). To ascertain if IL23A is indeed secreted, we first measured the accumulation of intracellular IL23A in RUNX3-expressing AGS following the inhibition of protein export by brefeldin A (BFA). This revealed a time-dependent increase in intracellular IL23A protein that peaked within 9 hr of exposure to TNF-α/H. pylori (Figure 4C). Moreover, a much higher level of intracellular IL23A is accumulated in RUNX3-expressing cells, compared with those of the control samples, underscoring the importance of RUNX3 (Figure 4D). Notably, IL23A accumulation is observed even in unstimulated RUNX3-expressing cells, indicating the exportation of IL23A in resting AGS cells (Figure 4D). To demonstrate the secretion of IL23A, supernatant of AGS cells transduced with a lentivirus encoding FLAG-tagged recombinant IL23A was subjected to immunoprecipitation by anti-FLAG antibodies. This resulted in the enrichment of tagged IL23A from the supernatant of transduced AGS cells (Figure 4E). Together, these data provide evidence for the secretion of IL23A by gastric epithelial cells, albeit in a form not recognized by canonical IL-23 ELISA kits.
Figure 4. Epithelial IL23A Is Secreted in a Form Distinct from Heterodimeric IL-23.
(A and B) Absence of heterodimeric IL-23 in gastric epithelial cell culture supernatants. (A) AGS cells were transduced with the indicated lentiviruses for 48 hr prior stimulation with TNF-α and/or H. pylori for 24 hr. Supernatants were subjected to sandwich ELISA for the quantification of IL-23 secretion. Representative data from two separate ELISA kits are presented. (B) Multiple gastric cancer cell lines were stimulated with TNF-α for 24 hr before culture supernatants were analyzed by sandwich ELISA specific for heterodimeric IL-23. Supernatant of THP-1 cells stimulated with LPS for 24 hr was included as a positive control (u.d., undetected).
(C) Intracellular accumulation of IL23A following the inhibition of protein export. AGS was transduced with Lenti-RUNX3 virus for 48 hr prior to TNF-α and H. pylori stimulation for the indicated times. Brefeldin A (BFA) was added to inhibit protein export 8 hr before each sample was harvested for western blotting analysis, using α-tubulin as control for protein loading.
(D) Intracellular IL23A is greatly induced by BFA in the presence of RUNX3. AGS cells were transduced with the indicated lentiviruses for 48 hr before treatment with TNF-α and H. pylori for 9 hr. BFA solution was added 8 hr prior to western blot analysis for the measurement of intracellular IL23A, RUNX3, and α-tubulin (loading control).
(E) IL23A is secreted by gastric epithelial cells. AGS cells transduced with the indicated lentiviruses were treated with TNF-α and H. pylori for 9 hr. Culture supernatant was subjected to immunoprecipitation by anti-FLAG agarose beads and analyzed by western blotting using IL23A-specific antibody.
(F) CD45/EpCAM expression profile of freshly isolated normal primary gastric cells. Cells were purified by FACS into CD45+ (immune) and EpCAM+CD45− (epithelial) fractions.
(G) Expression of IL23A, but not IL12B, in primary gastric epithelial cells. The expression of EpCAM, CD45, IL23A, and IL12B transcripts was measured in CD45+ and EpCAM+CD45− cells purified from human gastric tissues by qRT-PCR. Normalized values of each gene are expressed as a fraction of the sample with the higher expression.
Lastly, human primary gastric epithelial cells harvested from nonmalignant tissues adjacent to resected primary gastric tumors were analyzed for IL23A expression. The recovered cells were subjected to fluorescence-activated cell sorting (FACS), where the CD45+ leukocytic and CD45− EpCAM+ epithelial cell fractions were enriched (Figure 4F). The expression of IL23A was clearly detected by qRT-PCR in the purified epithelial fraction (Figure 4G). This was unlikely to be caused by contaminating leukocytes, since CD45 and IL12B were undetectable in this fraction. These observations in primary cells are consistent with those made in gastric carcinoma lines, confirming that IL23A is expressed independent of IL12B in human gastric epithelial cells.
DISCUSSION
The well-established link between chronic inflammation and cancer is particularly relevant in the gastrointestinal tract, where microbial contacts are frequent (Fox and Wang, 2007; Round and Mazmanian, 2009). In addition to being an important physical barrier, the epithelial cells in the gastrointestinal tract also create an immunological barrier that regulates the interactions of the luminal microbes with the mucosal immune system (Round and Mazmanian, 2009). In the stomach, the infection and colonization by H. pylori and the resultant injury to the epithelial lining are causally linked with chronic gastric inflammation and cancer (Fox and Wang, 2007). Among its highly adapted strategies, H. pylori delivers into the host cells bacterial peptidoglycan (iE-DAP) and cytotoxins like CagA to interfere and modify host-cell physiology (Odenbreit et al., 2000). These lead to changes such as altered epithelial cell-cell adhesion that facilitate colonization (Gebert et al., 2003). Concurrently, gastric epithelial cells in sensing these perturbations would mount an innate immune response, which includes the secretion of bactericidal peptides, chemokines, and proinflammatory cytokines to initiate inflammation (Censini et al., 1996). Indeed, while persistent and chronic inflammation promotes a vicious cycle of tissue damage and carcinogenesis, an effective acute inflammation is key to the eradication of pathogens (Fox and Wang, 2007; Round and Mazmanian, 2009).
This paradox is further reflected in the seemingly conflicting roles of the NF-κB pathway in immunity and disease. Despite its well-known contribution to chronic inflammation, autoimmunity, and cancer, recent studies have revealed that its epithelial function is unexpectedly protective (Nenci et al., 2007; Shibata et al., 2010; Zaph et al., 2007). This is attributed in part to NF-κB’s regulation of cytokines and chemokines, crucial for communication between epithelial cells and leukocytes. Of particular relevance, epithelial-specific targeting of IKK-β in the stomach resulted in defective clearance of H. pylori (Shibata et al., 2010).
Previous studies have shown that RUNX3 exerts its tumor- suppressor effects by modulating epithelial cell growth and maintaining epithelial integrity and barrier function (Chuang et al., 2013). The present study reports the transcriptional regulation of IL23A by RUNX3 in gastric epithelial cells, which is requisite for IL23A induction by the TNF-α/NF-κB pathway and H. pylori. This implicates IL23A as part of a NF-κB-driven antibacterial response mounted by gastric epithelial cells, as well as a role for RUNX3 in epithelial innate immunity. Moreover, the involvement of RUNX3 appears highly selective, as it had no effect on other IL-12 cytokine family members, nor did it augment the induction of IL8 by H. pylori.
IL23A mediates its known biological functions by dimerizing with IL12B to form IL-23, which is a potent proinflammatory cytokine essential for the functions of the IL-17-producing T helper cells (Th17) (Langrish et al., 2005). Reminiscent of the IKKβ knockout mouse phenotype, while IL23A null mice were protected against infection-induced inflammation, they succumbed to infection due to defective clearance, hence demonstrating a protective function for IL23A (Mangan et al., 2006). Although the primary source cells of the heterodimeric IL-23 are dendritic cells and macrophages, IL23A has been detected in other cell types, including keratinocytes, intestinal epithelial cells, and most recently gastric epithelial cells (Al-Sammak et al., 2013; Ciccia et al., 2009; Piskin et al., 2006). In this study, we observed the highly regulated expression of IL23A transcript and protein in a wide range of untransformed or transformed gastric epithelial cells. The secretion of IL23A by this cell type is supported first by the strong intracellular accumulation of IL23A following BFA treatment (Figure 4D), and second by the detection of tagged IL23A in culture supernatant by immunoprecipitation (Figure 4E). However, heterodimeric IL-23 (IL23A/IL12B) could not be detected by two independent sandwich ELISA kits, including one that employs distinct IL23A antibodies for capture and detection (data not shown). As previous studies have established that IL23A secretion is dependent on heterodimerization (Hunter, 2005; Oppmann et al., 2000), our data are consistent with the secretion of a noncanonical IL23A complex by gastric epithelial cells, in which IL23A’s interaction with a yet-unknown partner has masked epitopes utilized by existing ELISA kits.
Adding to the intrigue, RUNX proteins are also expressed in myeloid lineages, including dendritic cells and macrophages (Puig-Kröger and Corbí, 2006). Indeed, we observed the direct binding and transcriptional activation of IL23A promoter by RUNX3 and RUNX1 in ChIP and reporter assays using the human myeloid cell lines THP-1, U937, and K562 (Figures S1E and S3E). These data suggest that RUNX proteins may contribute to the production of distinct forms of IL23A via more than one cell type in the gastric mucosal microenvironment during inflammation and infection.
In summary, the current study reports the direct regulation of IL23A by RUNX3 in gastric epithelial cells, where its expression and secretion is independent of IL12B. The involvement of RUNX3 appears particularly important during inflammation and infection, as it strongly augmented the induction of IL23A by TNF-α/NF-κB and H. pylori. The identification of this pathway prompts further investigation into the participation of RUNX3 in the innate immunity of gastric epithelial cells and its contribution to pathogen clearance and mucosal homeostasis in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments
The human gastric lines AGS, KATOIII, MKN7, MKN28, MKN45, SNU16, NCI-N87, SNU719, and HFE-145 and the monocytic cell line THP-1 (Tohoku Hospital Paediatrics-1) were maintained in RPMI 1640 medium with 10% fetal bovine serum (FBS), penicillin/streptomycin, and 2 mM L-glutamine under standard tissue culture conditions. Cells were treated with 50 ng/ml of IFN-γ; 10 ng/ml of TNF-α, IL-1α, IL-1β, or IL-6; 5 ng/ml of TGF-β (Peprotech); 1 μg/ml of LPS, Zymosan, Pam3CSK4, Poly I:C, R-848; 100 μg/ml of Flagellin, 10 μg/ml of C12-iE-DAP; or 50 μg/ml of N-glycol MDP (InvivoGen) for 6 hr prior to analyses.
Reporter Gene Assay
The firefly luciferase reporter vector of IL23A-1200 and its variants were constructed as described in Supplemental Experimental Procedures. These reporter constructs were transfected into KATOIII, AGS, SNU16, K562, and U937 cells together with a variant of phRL-SV40 (Renilla luciferase) (Promega) and control (empty), RUNX3, or RUNX1 expression constructs using Lipofectamine 2000 reagent (Invitrogen) or FuGENE 6 (Promega) for 24–48 hr. Firefly luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega) and normalized against the corresponding Renilla luciferase values.
ChIP Analysis
AGS and HFE-145 cells transduced with Lenti-RUNX3 lentivirus and LPS-induced THP-1 cells were crosslinked in formaldehyde (1%) and sonicated to generate chromatin fragments of ~500 bp. ChIP assays were performed with the ChIP Assay Kit (Upstate Biotechnology) using polyclonal (AML2; Active Motif) and monoclonal (R3-5G4 and R3-6E9; MBL) antibodies against RUNX3, histone H3K9ac antibody (Abcam), and nonspecific rabbit or mouse immunoglobulin G (IgG) (Santa Cruz Biotechnology). The recovered DNA fragments were analyzed by quantitative PCR (qPCR) using specific oligo-nucleotide primers (refer to Table S1).
Gene Knockdown by RNAi and Overexpression by Lentiviruses
RNAi-mediated knockdown was accomplished by transfecting 0.2 nmol of gene-specific siRNA or nontargeting control siRNA (Thermo Scientific) using jetPRIME (Polyplus-transfection) on HFE-145 cells. For expression of exogenous RUNX3, RUNX3R178Q, and C-terminally tagged IL23A-FLAG, lentiviruses were prepared as previously described (Voon et al., 2012). For infection, cells were incubated with virus-containing supernatants for 24 hr and cultured with normal culture medium for another 24 hr.
qRT-PCR
Total RNA was extracted using the QIAGEN RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. cDNA was synthesized with the Omni-script RT kit (QIAGEN) as described by the accompanying protocol. qPCR was performed with the Universal PCR Master Mix (Applied Biosystems) using gene-specific oligonucleotide primers for SYBR Green-based measurements or TaqMan probes (Applied Biosystems) listed in Table S2. Data were analyzed on an ABI Prism 7500 real-time PCR system (Applied Biosystems). All gene expression data were normalized against GAPDH levels.
Protein Isolation and Western Blotting
Equal quantities of whole-cell lysates were resolved in SDS-polyacrylamide gel and immunoblotted with the following antibodies at their respective dilutions: anti-RUNX3 (1:1,500; R3-5G4), anti-RUNX1 (1:800; R1-1H9), anti-IL23A (1:250; eBio473P19, eBioscience), anti-α-tubulin (1:10,000; DM1A, Sigma-Aldrich), and anti-mouse IgG-horseradish peroxidase (1:5,000; Santa Cruz). To inhibit protein secretion, cells were treated with 1X BFA solution (eBioscience) 8 hr prior to harvest.
Preparation of H. pylori
Isogenic wild-type (NCTC11637) and CagA-defective (ΔCagA) strains of H. pylori are kind gifts from Dr. Hatakeyama (University of Tokyo, Japan). These are cultivated on Trypticase soy agars with 5% sheep blood (BD Biosciences) at 37°C in a humidified and microaerophilic atmosphere to form colonies. Broth cultures were prepared by inoculating colonies into Brucella broth (Sigma-Aldrich) with 10% FBS for 24 hr. Gastric cancer cell lines were infected with broth-cultured H. pylori at 100 MOI for indicated periods. For inhibition of the SHP-2/ERK pathway, cells were pretreated with 50 μM of SHP-2 inhibitor, NSC87877 (R&D Systems) for 3 hr prior to infection with H. pylori.
ELISA
For measurement of IL-23 in the supernatants harvested from cultured cells, ELISA was performed using the Human IL-23 Ready-SET-Go! kit (eBioscience) and the Human IL23A/p19 ELISA kit (Creative Biomart) as per the manufacturer’s instructions. To induce the secretion of IL-23, gastric epithelial cell lines and the monocytic cell line THP-1 were stimulated with TNF-α, H. pylori, or LPS for 24 hr.
Immunoprecipitation of FLAG Fusion IL23A
To enrich the FLAG-tagged IL23A in supernatant of AGS cells, immunoprecipitation (IP) was performed using mouse anti-FLAG M2 affinity gel (Sigma-Aldrich) following the manufacturer’s protocol. Supernatant was prepared from AGS cells transduced with lentivirus containing IL23A-FLAG construct. After 48 hr, cells were stimulated with TNF-α (10 ng/ml) and H. pylori (MOI100) for 9 hr prior harvesting the supernatant for the IP experiment.
Purification of Human Primary Gastric Epithelial Cells
All patient tissue samples were collected with informed patient consent from National University Hospital, Singapore, according to institutional review board guidelines. Resected gastric tissues were washed, minced, and incubated in 0.1% dithiothreitol/PBS to remove mucus. These were then digested with 100 U/mL hyaluronidase, 300 U/mL collagenase type II, 1 ng/ml hydrocortisone (Sigma-Aldrich), and 5 ng/mL insulin (Invitrogen) for 2 hr in Dulbecco’s modified Eagle’s medium/F12 (Invitrogen). Following washes and the removal of clumped and dead cells, cells were resuspended in 2% BSA/Hank’s balanced salt solution and stained with EpCAM-APC (Miltenyi Biotec) and CD45-FITC (Biolegend) antibodies and fractionated on a FACS Aria (BD Biosciences).
Statistical Analysis
All data are presented as mean ± SEM. The Student’s t test was utilized in comparison of two data sets, where p values of <0.05 are considered significant.
Supplementary Material
Acknowledgments
The authors acknowledge Masanori Hatakeyama, Ayumi Yamada, Kia Joo Puan, Vinay Tergaonkar, and Jimmy So for their generous provision of reagents and materials; and Qianqiao Tang, Cheong Kin Cheng, Herbert Schwarz, and Burkhard Becher for technical assistance and helpful discussion. This research is supported by the Translational and Clinical Research Flagship Programme (http://www.nmrc.gov.sg), the National Research Foundation Singapore (http://www.nrf.gov.sg) and the Singapore Ministry of Education, under its Research Centres of Excellence Initiative. Y.T.H. was a recipient of a National University of Singapore Graduate School for Integrative Sciences and Engineering scholarship.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.06.003.
AUTHOR CONTRIBUTIONS
Y.T.H. and D.C.-C.V. contributed equally to this manuscript. D.C.-C.V., Y.T.H., and Y.I. designed the study. Y.T.H. and D.C.-C.V. performed most of the experiments and analyzed the data. J.K.W.K. and H.W. assisted in the identification and validation studies. W.M.L. and S.L.C. contributed to the analyses in primary gastric cells. H.A. provided the HFE-145 line. D.C.-C.V. and Y.T.H. wrote the manuscript. Y.I. funded this work.
References
- Al-Sammak F, Kalinski T, Weinert S, Link A, Wex T, Malfertheiner P. Gastric epithelial expression of IL-12 cytokine family in Helicobacter pylori infection in human: is it head or tail of the coin? PLoS ONE. 2013;8:e75192. doi: 10.1371/journal.pone.0075192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AS. Regulation of cell death and autophagy by IKK and NF-κB: critical mechanisms in immune function and cancer. Immunol Rev. 2012;246:327–345. doi: 10.1111/j.1600-065X.2012.01095.x. [DOI] [PubMed] [Google Scholar]
- Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS, Ito K, Ito Y. RUNX family: regulation and diversification of roles through interacting proteins. Int J Cancer. 2013;132:1260–1271. doi: 10.1002/ijc.27964. [DOI] [PubMed] [Google Scholar]
- Ciccia F, Bombardieri M, Principato A, Giardina A, Tripodo C, Porcasi R, Peralta S, Franco V, Giardina E, Craxi A, et al. Overexpression of interleukin-23, but not interleukin-17, as an immunologic signature of subclinical intestinal inflammation in ankylosing spondylitis. Arthritis Rheum. 2009;60:955–965. doi: 10.1002/art.24389. [DOI] [PubMed] [Google Scholar]
- Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Wang TC. Inflammation, atrophy, and gastric cancer. J Clin Invest. 2007;117:60–69. doi: 10.1172/JCI30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301:1099–1102. doi: 10.1126/science.1086871. [DOI] [PubMed] [Google Scholar]
- Hunter CA. New IL-12-family members: IL-23 and IL-27, cytokines with divergent functions. Nat Rev Immunol. 2005;5:521–531. doi: 10.1038/nri1648. [DOI] [PubMed] [Google Scholar]
- Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res. 2005;65:7743–7750. doi: 10.1158/0008-5472.CAN-05-0743. [DOI] [PubMed] [Google Scholar]
- Ito K, Chuang LS, Ito T, Chang TL, Fukamachi H, Salto-Tellez M, Ito Y. Loss of Runx3 is a key event in inducing precancerous state of the stomach. Gastroenterology. 2011;140:1536–1546.e8. doi: 10.1053/j.gastro.2011.01.043. [DOI] [PubMed] [Google Scholar]
- Jobin C, Sartor RB. The I kappa B/NF-kappa B system: a key determinant of mucosalinflammation and protection. Am J Physiol Cell Physiol. 2000;278:C451–C462. doi: 10.1152/ajpcell.2000.278.3.C451. [DOI] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D, Negreanu V, Lotem J, Bone KR, Brenner O, Leshkowitz D, Groner Y. Transcription factor Runx3 regulates interleukin-15-dependent natural killer cell activation. Mol Cell Biol. 2014;34:1158–1169. doi: 10.1128/MCB.01202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue Ki, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109:113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- Nenci A, Becker C, Wullaert A, Gareus R, van Loo G, Danese S, Huth M, Nikolaev A, Neufert C, Madison B, et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- Odenbreit S, Püls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- Oppmann B, Lesley R, Blom B, Timans JC, Xu Y, Hunte B, Vega F, Yu N, Wang J, Singh K, et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- Piskin G, Sylva-Steenland RM, Bos JD, Teunissen MB. In vitro and in situ expression of IL-23 by keratinocytes in healthy skin and psoriasis lesions: enhanced expression in psoriatic skin. J Immunol. 2006;176:1908–1915. doi: 10.4049/jimmunol.176.3.1908. [DOI] [PubMed] [Google Scholar]
- Puig-Kröger A, Corbí A. RUNX3: a new player in myeloid gene expression and immune response. J Cell Biochem. 2006;98:744–756. doi: 10.1002/jcb.20813. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata W, Takaishi S, Muthupalani S, Pritchard DM, Whary MT, Rogers AB, Fox JG, Betz KS, Kaestner KH, Karin M, et al. Conditional deletion of IkappaB-kinase-beta accelerates helicobacter-dependent gastric apoptosis, proliferation, and preneoplasia. Gastroenterology. 2010;138:1022–1034.e1. 10. doi: 10.1053/j.gastro.2009.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon DC, Wang H, Koo JK, Nguyen TA, Hor YT, Chu YS, Ito K, Fukamachi H, Chan SL, Thiery JP, Ito Y. Runx3 protects gastric epithelial cells against epithelial-mesenchymal transition-induced cellular plasticity and tumorigenicity. Stem Cells. 2012;30:2088–2099. doi: 10.1002/stem.1183. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Sugai M, Nambu Y, Osato M, Hayashi T, Kawaguchi M, Komori T, Ito Y, Shimizu A. Requirement for Runx proteins in IgA class switching acting downstream of TGF-beta 1 and retinoic acid signaling. J Immunol. 2010;184:2785–2792. doi: 10.4049/jimmunol.0901823. [DOI] [PubMed] [Google Scholar]
- Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, Du Y, Yost EA, Gruber AD, May MJ, Greten FR, et al. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.