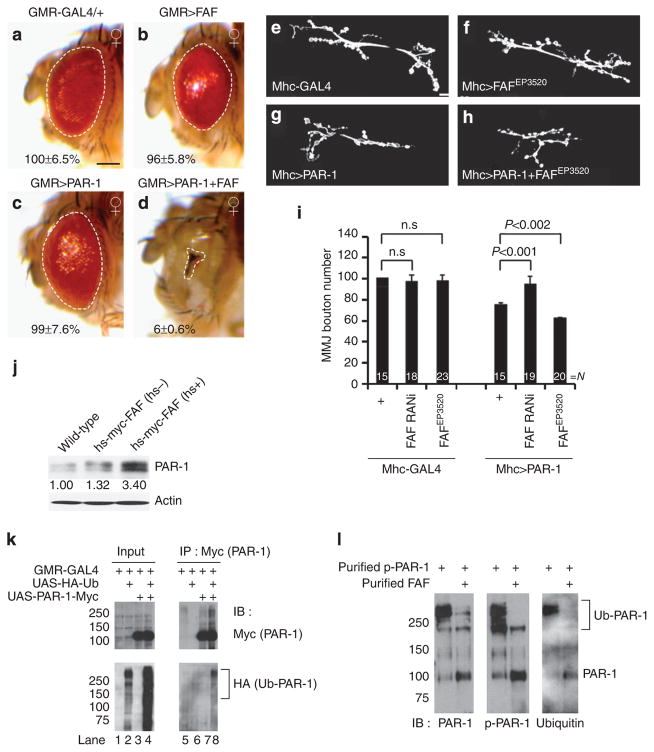
Figure 1. FAF positively regulates PAR-1 activity and protein stability.
(a–d) Genetic interaction between PAR-1 and FAF in the fly retina. All flies were grown at 25 °C. Images of female flies are shown. The genotypes are: GMR-Gal4/ +control (a), GMR-Gal4>FAFEP381 (b), GMR-Gal4>UAS-PAR-1 (c) and GMR-Gal4>UAS-PAR-1 +FAFEP381 (d) (n =16, 17, 16 and 16 animals, respectively). Statistically significant differences are P<0.001 (GMR-Gal4>UAS-PAR-1, GMR-Gal4>UAS-PAR-1 +FAFEP381) as determined by Student’s t-test. Experiments were performed in triplicate. Dashed lines outline the eye contour. Values represent areas of retinal surface normalized with GMR-Gal4/ +control. Scale bar (a–d), 100 μm. (e–h) Representative NMJ terminals of the indicated genotypes revelled by anti-horseradish peroxidase (HRP) immunostaining. The genotypes are Mhc-Gal4/ +control (e), Mhc-Gal4>FAFEP3520 (f), Mhc-Gal4>UAS-PAR-1 (g) and Mhc-Gal4>UAS-PAR-1 +FAFEP3520 (h). Scale bar (e–h), 10 μm (i) Quantification of the total number of boutons per muscle area on muscle 6/7 of A3 in the indicated genotypes. N indicates the number of animals analysed. The error bars represent means±s.e.m. P-values were determined using two-tailed Student’s t-test for each comparison. Experiments were performed in triplicate. (j) Western blot analysis of endogenous PAR-1 protein level after FAF induction from hs-Myc-FAF. Actin serves as a loading control. Values represent PAR-1 levels normalized with wild-type control in three independent experiments. (k) Western blot analysis showing in vivo ubiquitination of PAR-1 in animals co-expressing PAR-1-Myc and HA-Ub (lane 8). (l) In vitro deubiquitination assay using affinity-purified, HA-Ub-modified phsopho-PAR-1 as the substrate, and affinity-purified FAF as the enzyme. Note the decrease of poly-ubiquitinated PAR-1 and the corresponding increase of non-ubiquitinated or mono-ubiquitinated PAR-1 after FAF treatment. Brackets indicate ubiquitinated PAR-1 (Ub-PAR-1) in (k,l.). IB, immunoblot; n.s., not significant.