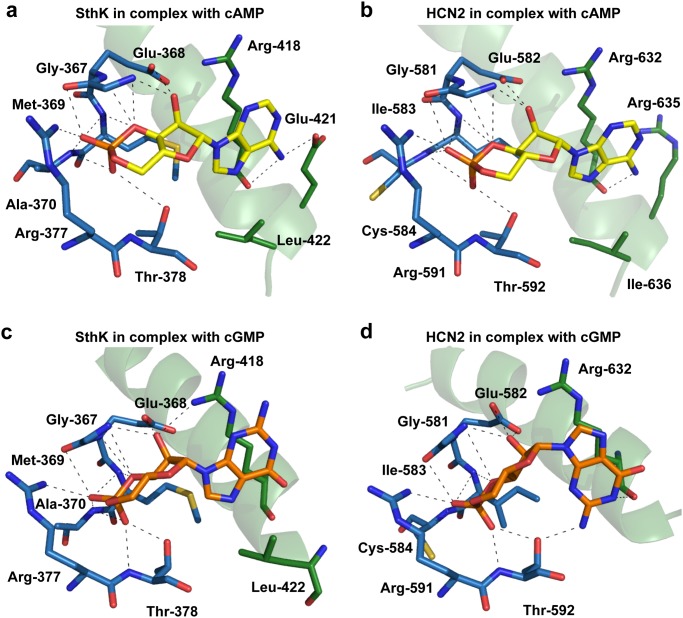
Fig 3. Molecular determinants of ligand recognition in the SthK-Cterm in complex with cAMP or cGMP.
a-b) Comparison of the amino acids in the ligand binding site involved in recognition of cAMP in SthK (left) and HCN2 (right). Amino acids of the β-roll are shown in blue sticks, residues of the C-helix in green sticks. The C-helix is shown in cartoon representation. The cAMP molecule is shown in yellow. Dashed lines represent hydrogen bonds or salt bridges. c-d) Comparison of the cGMP bound SthK-Cterm with the cGMP bound structure of HCN2 illustrates that the cGMP molecule binds in the anti-conformation, whereas previously published structures showed cGMP bound in its syn-conformation. Locked in the anti-conformation, the phosphoribose part of cGMP is able to establish the same interaction with SthK-CNBD as cAMP. However, the anti-conformation appears to inhibit interactions between the purine ring from cGMP and the C-helix from the SthK-CNBD. Most likely, the absence of these interactions explains why binding of cGMP to SthK does not provide the required energy for the opening conformational changes.