Abstract
The E-proteins and Id-proteins are, respectively, the positive and negative heterodimer partners for the basic-helix-loop-helix protein family, and as such contribute to a remarkably large number of cell fate decisions. E-proteins and Id-proteins also function to inhibit or promote cell proliferation and cancer. Using a genetic modifier screen in Drosophila, we show that the Id-protein Extramacrochaetae enables growth by suppressing activation of the Salvador-Warts-Hippo pathway of tumor suppressors, activation that requires transcriptional activation of the expanded gene by the E-protein Daughterless. Daughterless protein binds to an intronic enhancer in the expanded gene, both activating the SWH pathway independently of the transmembrane protein Crumbs, and bypassing the negative feedback regulation that targets the same expanded enhancer. Thus the Salvador-Warts-Hippo pathway has a cell-autonomous function to prevent inappropriate differentiation due to transcription factor imbalance, and monitors the intrinsic developmental status of progenitor cells, distinct from any responses to cell-cell interactions.
Introduction
The coordination of differentiation with growth and proliferation is important so that organs develop with proper organization and size. Failures of this coordination may cause organ malformations or tumors. We describe how certain defective cells are recognized and prevented from causing neural defects by the Salvador-Warts-Hippo (SWH) pathway.
Transcription factors of the basic helix-loop-helix (bHLH) family contribute to a remarkably large number of cell fate decisions. E-proteins are bHLH transcription factors that bind DNA at E-box (CANNTG) consensus sequences. E proteins appear to be expressed in every cell, and heterodimerize with tissue-specific bHLH proteins to play specific roles in differentiation (Massari and Murre, 2000). Drosophila has a single E-protein, Daughterless (Da), which in combination with heterodimer partners is required for neurogenesis, sex determination, and mesoderm development (Murre et al., 1989; Goulding et al., 2000; Huang et al., 2000; Massari and Murre, 2000). The 4 mammalian E-proteins E12 and E47 (also known as TCF3), E2-2 (TCF4), and HEB (TCF12) play crucial roles in the regulation of commitment, cell growth and differentiation in lymphocytes, muscle cells, neurons, and other cells (Massari and Murre, 2000; Slattery et al., 2008; Kee, 2009).
By contrast to E-proteins, Id-proteins prevent DNA binding and function when they heterodimerize with either E-proteins or tissue-specific bHLH proteins like the muscle specific protein MyoD (Benezra et al., 1990; Lassar et al., 1991) or proneural genes of the Achaete-Scute Complex (AS-C) (Van Doren et al., 1991). Levels of the only Drosophila Id-protein, Extramacrochaetae (Emc), set thresholds for differentiation in response to positively-acting bHLH heterodimers. Accordingly, hypomorphic mutations that reduce emc function lead to ectopic neural differentiation, whereas null alleles are lethal to the embryo (Van Doren et al., 1992; Cubas and Modolell, 1992).
It has been known for many years that clones of cells completely lacking emc cannot easily be recovered in growing imaginal disc tissues (Garcia Alonso and Garcia-Bellido, 1988; de Celis et al., 1995). This implies a role for emc in cell proliferation or survival as well as in differentiation, but the mechanism of this growth contribution was not known. Mammalian Id genes are also growth regulators and can act as proto-oncogenes (Iavarone et al., 1994; Lasorella et al., 1996; Norton, 2000; Hasskarl and Munger, 2002; Perk et al., 2005).
The Salvador-Warts-Hippo (SWH) pathway has emerged as a new growth pathway, largely conserved between Drosophila and mammals (Bossuyt et al., 2014). The core components are the cytoplasmic Hpo and Wts kinases that, together with their accessory proteins Sav and Mats, phosphorylate and retain Yorkie (Yki)/YAP in the cytoplasm (Tapon et al., 2002; Harvey et al., 2003; Jia et al., 2003; Pantalacci et al., 2003; Udan et al., 2003; Wu et al., 2003; Huang et al., 2005; Lai et al., 2005). Yki/YAP is a transcriptional coactivator that lacks DNA-binding activity but interacts with proteins such as Scalloped/TEAD family transcription factors to mediate Yki/YAP-induced gene expression (Wu et al., 2008; Goulev et al., 2008; Zhao et al., 2008; Zhang et al., 2008). Yki/YAP can promote cell proliferation, particularly in progenitors and stem cells. Deregulation of Yki/YAP promotes tissue overgrowth and tumor formation, whereas Yki/Yap activity is required for normal growth and for tissue regeneration (Ramos and Camargo, 2012). Mutations in the upstream Hpo pathway genes lead to overgrowth phenotypes in Drosophila and are implicated in various human cancers (Pan, 2010; Grusche et al., 2010; Zhao et al., 2011; McCaffrey and Macara, 2011; Genevet and Tapon, 2011; Staley and Irvine, 2012; Schroeder and Halder, 2012).
Hpo and Wts activity depend on more upstream proteins that suggest the SWH pathway is regulated by cell-cell interactions. These include the apical membrane protein Crumbs (Crb), the protocadherin Fat/Fat4, the FERM domain proteins Expanded (Ex)/FRMD6, Merlin (Mer)/NF2 and Kibra (Hamaratoglu et al., 2006; Willecke et al., 2006; Feng and Irvine, 2007; Baumgartner et al., 2010; Ling et al., 2010; Robinson et al., 2010; Yu et al., 2010; Chen et al., 2010; Genevet et al., 2010). Fat can interact with another protocadherin, Dachsous, expressed on neighboring cells, providing a potential cell-cell signaling pathway influencing growth (Cho et al., 2006; Rogulja et al., 2008; Willecke et al., 2008; Schwank et al., 2011). Crb seems to regulate SWH through homophilic interactions (Ling et al., 2010; Robinson et al., 2010; Chen et al., 2010). Evidence is increasing that SWH activity may be influenced mechanically, perhaps through the actin cytoskeleton as well as through cell junctions (Boggiano and Fehon, 2012; Schroeder and Halder, 2012).
The physiological roles of the SWH pathway are not yet fully understood (Lawrence and Casal, 2013). Although it is hypothesized that the SWH pathway controls organ growth, direct evidence is lacking that normal growth terminates due to an increase in SWH activity. On the other hand, loss of SWH regulation may underlie tissue expansion that occurs in wound healing and regeneration (Cai et al., 2010; Karpowicz et al., 2010; Shaw et al., 2010; Zhao et al., 2011; Grusche et al., 2011; Barry et al., 2013), and in the expansion of the wing primordium during Drosophila development (Zecca and Struhl, 2010). While reduced SWH pathway activity is implicated in growth stimulation, evidence that SWH hyperactivity blocks growth and cell survival is mainly found in experimental situations (Jia et al., 2003; Udan et al., 2003; Bennett and Harvey, 2006; Dong et al., 2007; Tyler and Baker, 2007). The SWH pathway is thought to mediate contact inhibition in mammalian cells (Lallemand et al., 2003; Zhao et al., 2007) as well as the distinction between trophectoderm and inner cell mass in mouse embryogenesis (Nishioka et al., 2009). The core of the SWH pathway is also required for selection of rhodopsin expression in the postmitotic retina, another process that depends on cell-cell interactions (Jukam and Desplan, 2011).
In this study we report that unbalanced bHLH expression activates the SWH pathway through the direct transcriptional regulation of the ex gene by the E-protein Daughterless. Our findings indicate that SWH activity prevents emc mutant cells from causing inappropriate ectopic neurogenesis. Our findings therefore identify a role for the SWH pathway in the cell-autonomous recognition and elimination of certain mis-specified cells during development.
Results
Emc regulates growth through Da
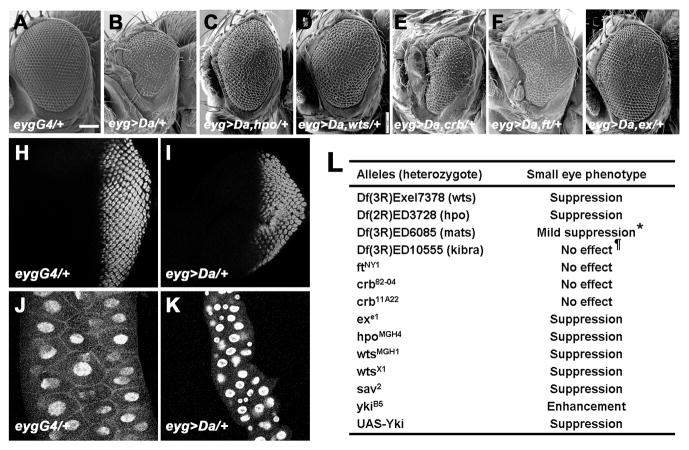
Like mammalian Id-proteins, the Drosophila Id-protein Extramacrochaetae (Emc) is a HLH protein lacking a basic domain whose heterodimers are unable to bind DNA (Benezra et al., 1990; Campuzano, 2001). The emc gene promotes cell growth and survival, and it is hard to recover clones of progenitor cells in imaginal discs that lack emc (Garcia Alonso and Garcia-Bellido, 1988; de Celis et al., 1995). Because Emc expression is regulated by the E-protein Daughterless (Da), and serves as a negative feedback regulator of daughterless expression and activity, emc null mutant cells express high levels of Da proteins that are responsible for their growth defect (Bhattacharya and Baker, 2011). Consistent with this finding, Da overexpression during growth under the control of eyg-Gal4 can reduce the size of adult organs and larval tissues including the eye and the salivary gland (Figures 1A–B and 1H–K, and Figure S1A–B). Therefore, excess Da reduced both diploid, mitotic growth of imaginal discs and endoreplicative growth of the salivary gland. As expected if eyg>da mimics the growth effects of emc loss, the small eye was further reduced by loss of one copy of the endogenous emc gene, but partially restored by loss of one copy of the endogenous da gene, and eye size was also reduced by RNA interference of emc by a dsRNA (Figure S1D–F and data not shown).
Figure 1. SWH Pathway Components Are Required for Excess Da-induced Growth and Survival Defect.
(A–G) Scanning electron micrographs of adult eyes. Scale bar, 100 μm. (A) Control eygCD-GAL4/+ flies. (B) eygCD-GAL4 driving UAS-Da flies (abbreviated as eyg>Da). (C) eyg>Da, hpoMGH4/+ flies. (D) eyg>Da, wtsX1/+ flies. (E) eyg>Da, crb82-04/+ flies. (F) eyg>Da, ftNY1/+ flies. (G) eygCD>Da, exe1/+ flies. (H–I) Third instar eye imaginal disc of eygCD-GAL4 (H) or eyg>da (I) staining for anti-Elav (red). (J–K) Third instar salivary gland of eygCD-GAL4 (J) or eyg>da (K) staining for DAPI (blue). (L) A subset of SWH pathway genes genetically interact with eyg>Da. *Results with mats were equivocal. The other independent mutant of mats, matse235, did not suppress eyg>Da. ¶Using TRiP RNA against kibra partially suppressed eyg>Da. See also Figure S1 and Tables S1–S2.
Da functions with the Salvador-Warts-Hippo pathway
To elucidate the mechanism by which Da affects cell survival and growth, we performed a dominant genetic modifier screen using a series of chromosomal deletions (Parks et al., 2004; Ryder et al., 2004; Roote and Russell, 2012; Cook et al., 2012) to identify loci that were dose-sensitive for growth effects of high Da (Table S1). The size of the eyg>da eye was modified by reduced gene dose of multiple cell death and cell cycle regulators, including head involution defective, grim, reaper, sickle, string, wee1, Rb, E2f1, Cyclin A, and Cyclin E, and was rescued by co-expression of the anti-apoptotic proteins baculovirus p35, Diap1, or dominant-negative Dronc (Table S2). Two of the regions with strong dominant suppression of Da-induced growth inhibition were cytological intervals 56D10-56E2 and 99F8-100A5, which uncovered hippo (hpo) and warts (wts) respectively. Multiple independent point mutants of hpo and wts were all then found to suppress the Da overexpression phenotype (Figures 1C–D). For comparison, the eygCD>da small eye phenotype was not modified by heterozygosity for ptenMGH1 (data not shown), a null allele of a negative regulator of the Insulin pathway, indicating that the genetic interaction with hpo and wts was not shared by all growth regulators. By contrast, when Da was over-expressed in the differentiating, post-mitotic eye using GMR-GAL4, the effects on differentiation that resulted were not modified by hpo or wts gene dose, showing that hpo and wts primarily affected Da function during growth (Figures S1G–H). This latter finding also argued that hpo and wts did not simply modify transcriptional activation by Gal4. Taken together, these results suggested that Da acted through Hpo and Wts, two components of the SWH pathway that coordinately regulates growth and survival.
To establish how much of the SWH pathway was implicated in the growth response to high Da, point mutations for components that were not uncovered by the deficiency collections were tested. Growth suppression in Da-overexpressing eyes also depended on the gene dose of Sav, Yki, and the adaptor protein ex (Figures 1G and 1L). By contrast, eyg>da was not modified by dosage of the transmembrane protein genes ft or crb (Figures 1E–F).
The SWH pathway is epistatic to Da in growth control
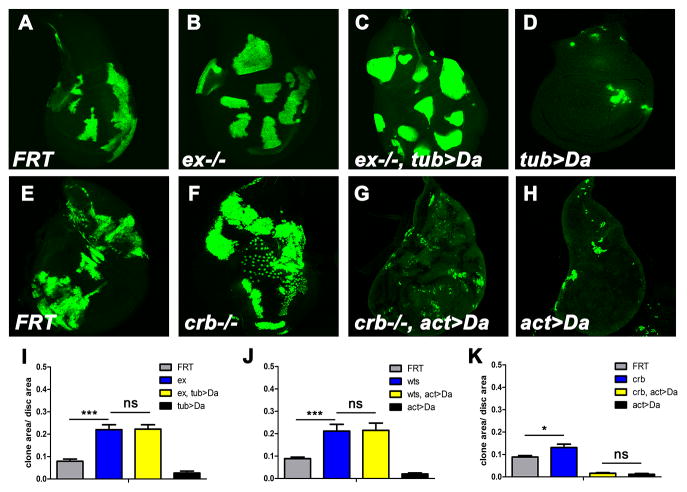
To establish how quantitatively growth suppression by Da was dependent on the SWH pathway, Da was over-expressed specifically in clones of cells that also lacked particular SWH components. Mosaic analyses with a repressible cell marker (MARCM) (Lee and Luo, 1999) was used to generate null clones of ex that also over-express Da. The sizes of exe1; tub>Da clones were indistinguishable from exe1 clones (Figures 2A–D and 2I), indicating that Da over-expression (tub>Da) did not inhibit clone growth in the absence of ex. Similar results were found in wtsX1 mutant clones (Figures 2J and S2A–D). Since Da protein level was not affected in wts clones (Figure S2J), these findings indicated that wts and ex were epistatic to high Da, and required for high Da to affect growth. By contrast, clones overexpressing Da and mutant for crb rarely survived in imaginal discs, indicating that Da over-expression continued to inhibit growth in the absence of crb (Figures 2E–H and 2K). High Da did not affect growth in the absence of ft (Figures S2E–I), perhaps because Ft can affect Wts independently of parts of the pathway (Cho et al., 2006). Together, the results indicated that Da required the core SWH pathway to inhibit growth, but did not require the transmembrane protein Crb.
Figure 2. Da Over-expression acts upstream of ex.
(A–D) Third instar wing imaginal discs with positively marked MARCM clones stained for GFP (green, to mark the clones). wild-type control (A), exe1 (B), exe1, tub>Da (C), and tub>Da (D). Although clones of exe1 or exe1, tub>Da grew well, tub>Da clones were rarely recovered in the disc proper. (E–H) Third instar wing imaginal discs with wild-type control (E), crb82-04 (F), crb82-04, act>Da (G), and act>Da (H). Loss of crb did not affect clone loss when Da was over-expressed. (I–K) Quantification results of A–D, E–H and Figure S2A–D, respectively. 10 discs of each genotype are analyzed. Areas of clones are normalized to the total wing disc area. Mean±SEM is shown, significance assessed with Student’s t-test; *** P<0.001; ** P<0.01; * P<0.05; NS, not significant. See also Figure S2.
Da regulates ex transcription
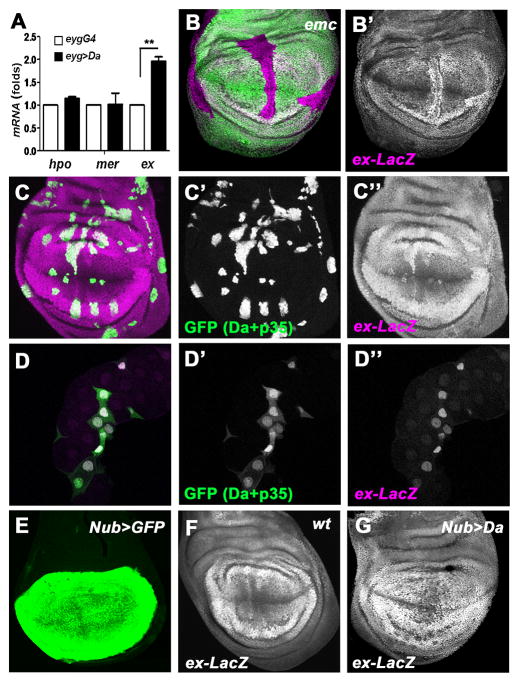
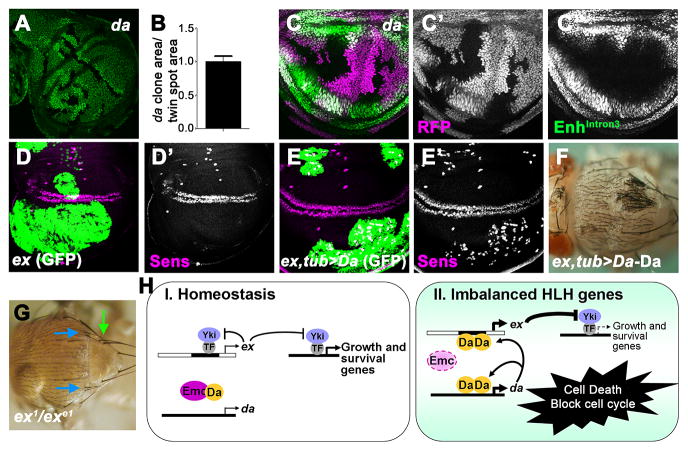
Since Da is a transcription factor, it might regulate transcription of SWH genes. Quantitative RT-PCR analysis was performed to identify potential targets. The mRNA levels of ex were increased in Da-overexpressing tissue when compared to wild-type, while hpo and mer mRNA levels did not show significant changes (Figure 3A). Since ex was also the most upstream SWH component that acted as a genetic modifier of Da, the transcription of ex was examined in situ using an enhancer trap line, ex-LacZ. When Da was over-expressed under Actin-Gal4 control using a Flp-out method, ex transcription was cell-autonomously elevated as indicated by the ex-LacZ reporter (Figures 3C–D). Similar results were seen in both diploid imaginal disc cells, and in polytene salivary gland cells that grow by endoreplication. The anti-apoptotic baculovirus p35 was co-expressed with Da in these experiments to preserve cells over-expressing Da. ex-LacZ was not affected in otherwise wild type cells expressing p35 (Figures S3A–B). To investigate ex transcription in another way, the Nubbin-Gal4 driver (Nub-Gal4) was used to over-express Da. Nub-Gal4 is active specifically in the wing pouch (Figure 3E), and Nub>da clearly elevated ex transcription in most such cells as shown both by the ex-LacZ reporter and antibody staining for the Ex protein (Figures 3F–G; Figures S3D–E). Nub>da leads to cell death in the wing disc and a great reduction in adult wing size that is partially rescued by co-expression of the antiapoptotic protein baculovirus p35 (Figures S3F–L). Finally, the ex-LacZ reporter was also up-regulated in clones of emc mutant cells in the wing disc, confirming that elevated Da expression mimics the effects of mutating emc (Figure 3B). These findings indicate that high levels of Da activate ex transcription. Elevated ex transcription is known to be sufficient to activate the SWH pathway, epistatically to certain transmembrane receptors (Hamaratoglu et al., 2006; Tyler and Baker, 2007).
Figure 3. ex Is a Transcriptional Target of Da.
(A) A graph comparing hpo, mer and ex mRNA levels in wild type and Da-overexpressing salivary glands, as measured by qRT-PCR. Results represent mean±SEM (n=3). ** P<0.01. The mRNA levels of ex are increased 1.96±0.17 fold. (B) A wing disc containing emc mutant clones (GFP negative) stained for ex-lacZ reporter expression (magenta). Transcriptional activity of the ex gene is reported by detecting β-galactosidase expression. The increased levels of ex-lacZ in emc mutant cells are most obvious in the wing pouch. (C) Wing imaginal discs containing Da-overexpressing clones (ActGal4>GFP+Da+P35, GFP positive, green) stained for ex-lacZ reporter expression (magenta). The anti-apoptotic baculovirus p35 was co-expressed with Da in these experiments to preserve cells over-expressing Da. The ex-LacZ reporter was elevated by high Da cell-autonomously. (D) Salivary glands containing Da-overexpressing clones (ActGal4>GFP+Da+P35, GFP positive, green) and stained for ex-lacZ reporter expression (magenta). Note ex-lacZ was elevated in Da-overexpressing clones. Salivary glands from Da-overexpression animals are often smaller, in addition to the cell-autonomous effect of Da-overexpression. (E) UAS-GFP under the control of Nub-GAL4 in the wing imaginal discs. Note GFP is detected in the wing pouch region. (F, G) Wing imaginal discs of wild type (F) and Nub>Da (G) containing ex-LacZ (magenta). Note the elevation of ex-LacZ reporter in Da-overexpressing wing pouch cells. See also Figure S3.
Identification of cis-regulatory elements for the ex gene
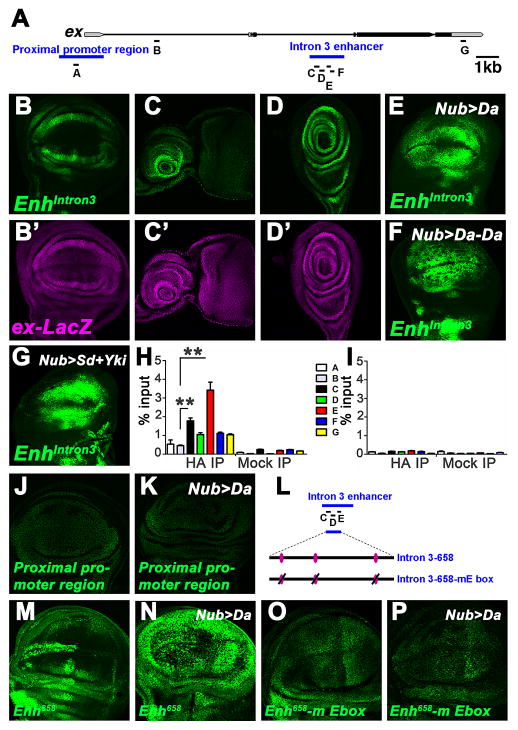
To determine whether ex was a direct transcriptional target of Da, we first sought the Da response element of the ex gene. A ChIP-chip database of early embryogenesis (MacArthur et al., 2009) reported modest association of Da protein with two regions. These putative Da-response elements were tested in vivo for enhancer activity (Figure 4A). The region within the third ex intron drove GFP reporter gene expression in patterns identical to ex-LacZ (Figures 4B–D). To determine whether Intron 3 enhancer (EnhIntron3) confers response to Da, Da was over-expressed in the wing pouch, eye disc or salivary glands. GFP expression was elevated in Da over-expressing cells (Figure 4E and not shown), indicating that EnhIntron3 is a Da-responsive enhancer. By contrast, the proximal promoter region did not respond to Da overexpression in wing imaginal discs (Figures 4J–K).
Figure 4. Characterization of ex Enhancers.
(A) Schematic representation of the ex locus. Exons are represented as black arrows, introns are represented by lines. Non-coding regions are shown as grey arrows. Blue bars label Proximal promoter region and Intron 3 enhancer (Enhintron3). Bars labeled A–G indicated regions amplified by PCR in ChIP experiments. (B–D) Expression patterns of Enh intron3-GFP and ex-LacZ in the wing (B, B′), eye (C, C′), or leg (D, D′) discs. (E–G) Wing discs of Nub>Da (E), Nub>Da-Da homodimer (F), and Nub>Sd+Yki (G) staining for Enh intron3-GFP reporter expression. Note the upregulation of Enh intron3-GFP levels in all these wing discs. (H) ChIP analysis of the EnhIntron3 enhancer. Anti-HA (HA IP) or IgG antibodies are used to precipitate chromatin from wing discs of Nub-Gal4>UAS-HA-Da. Quantitative PCR was done on regions A–G (see panel A). Graph shows the percentage of signal relative to input. Results represent mean±SEM (n=3). **P<0.01 (t-test). (I) Control HA ChIP. Anti-HA antibody is used to precipitate chromatin from Nub-Gal4 wing discs. (J) Wild-type wing disc staining for Proximal promoter region-GFP expression. (K) Wing disc of Nub>Da staining for Proximal promoter region-GFP expression. The Proximal promoter region-GFP expression was unaffected by Da over-expression. (L) Schematic representation of the Intron 3 enhancer. Bars labeled C, D, E indicate ChIP amplicons. Subfragment (658 bp) covering C to E regions contains 3 putative E-box elements (purple ovals). (M) Wild-type wing disc staining for Intron 3-658-GFP (abbreviated as Enh658) expression. Unlike the complete enhancer, this construct drives more GFP expression in cells at the anterior wing margin. (N) Wing disc of Nub>Da staining for Enh658 expression. Note the elevation of Enh 658-GFP levels throughout the Nub-Gal4 domain. (O) Intron 3-658-GFP loses expression with three mutated E-box elements (abbreviated as Enh658-mE box). (P) Enh658-mE box expression in Nub>Da wing disc. Note that the Enh658-mE box did not respond to Da.
To assess whether Da directly associates with EnhIntron3 in larval tissues, chromatin immunoprecipitation (ChIP) experiments were conducted with chromatin isolated from wing imaginal discs in which Nub-GAL4 drove HA-tagged Da. DNA amplified from chromatin immunoprecipitated with anti- HA was significantly enriched in regions C and E within the identified enhancer element (Figure 4H). Other parts of the enhancer showed less enrichment or were not enriched compared to negative controls (amplicon A near the transcription start site, B in the first intron, and G in the 3′ UTR). None of the regions was enriched when immunoprecipitated with control IgG antiserum, or in HA-immunoprecipitated chromatin prepared from wing discs of Nub-GAL4 flies (Figures 4H–I), although Nub-GAL4 chromatin could be precipitated with other antibodies (Figure S4F). Da-responsiveness was retained by a 658 bp fragment covering the regions with enriched Da association, that contains three E-box sequences that are putative binding sites for bHLH proteins (Figures 4L–N). Da-responsiveness was lost when these E-boxes were mutated (Figures 4L and 4O–P). Thus, Da protein is bound to the Da-responsive element of ex in wing discs in vivo, consistent with direct binding as the mechanism of activation by high Da levels.
Da affected ex expression in cells that are not known to express any bHLH heterodimer partners (Figures 3B–C and 4E). Although Da functions as a heterodimer with proneural bHLH proteins where they are expressed, and ectopic expression of the proneural proteins Achaete and Scute could activate the enhancer (Figures S4D–E). Da can also homodimerize and Da homodimers can bind to DNA (Murre et al., 1989; Jarman et al., 1993; Huang et al., 2000; Jafar-Nejad et al., 2003). Indeed, we found that EnhIntron3-GFP was upregulated in the wing imaginal epithelium when a covalent Da-Da homodimer was expressed (Figure 4F), indicating that SWH activity could be regulated by Da homodimers and that this can occur outside of proneural regions (Figures S4D, E). Covalent Da-Da dimer expression led to phenotypes similar to over-expression of Da monomer, but more severe (Fig. S4A–C and data not shown).
In addition to its essential role in SWH signaling, ex is also a transcriptional target of Yki. Since Yki is inhibited by SWH activity, regulated ex transcription constitutes a negative feedback loop for the SWH pathway, whereby diminished SWH activity disinhibits Yki, upregulating ex transcription and restoring SWH activity (Hamaratoglu et al., 2006). Since the expression pattern of EnhIntron3-GFP mimicked ex-lacZ expression in multiple imaginal discs even in the absence of Da over-expression, we determined whether EnhIntron3 also encoded the feedback of Yki on ex expression. Indeed, co-expressing Scalloped (Sd: the DNA-binding protein partner for Yki during wing development (Goulev et al., 2008; Zhang et al., 2008; Pan, 2010; Zhao et al., 2011) and Yki elevated EnhIntron3-GFP in the wing discs (Figure 4G). In sum, these data demonstrate that EnhIntron3 is a cis-regulatory element of ex regulated by the bHLH protein Da as well as by the SWH pathway through Sd/Yki.
Da over-expression reduces Yki activity
Our data show that Da requires SWH activity to affect growth (Figures 1, 2 and S2). If elevated ex expression and SWH activity is the relevant mechanism, then Yki target gene expression should be affected by Da. To test this, we monitored the miRNA bantam (ban) using the ban-GFP sensor (Brennecke et al., 2003). When Da was over-expressed, either clonally or using Nub-GAL4, a significant increase of ban-GFP sensor was revealed in wing discs (Figures 5A–C, S5B). Because ban destabilizes the ban-GFP sensor, this indicated reduction of ban miRNA levels and Yki activity in Da over-expressing cells. Another assay examined the four-joint enhancer trap (fj-lacZ) or diap1 reporter (diap1-lacZ) as readouts of Yki activity (Cho et al., 2006; Wu et al., 2008). Expression of fj-lacZ and diap1-lacZ were decreased in emc clones or Da overexpressing cells (Figures 5D, S5C, S5F), indicating reduced Yki activity when Da is upregulated. This effect of Da was dependent on ex (Figure S5E). Collectively, these data confirmed that high Da both activated ex transcription and reduced Yki activity, as expected if this is the mechanism of growth control by high Da.
Figure 5. High Da affects multiple SWH pathway genes.
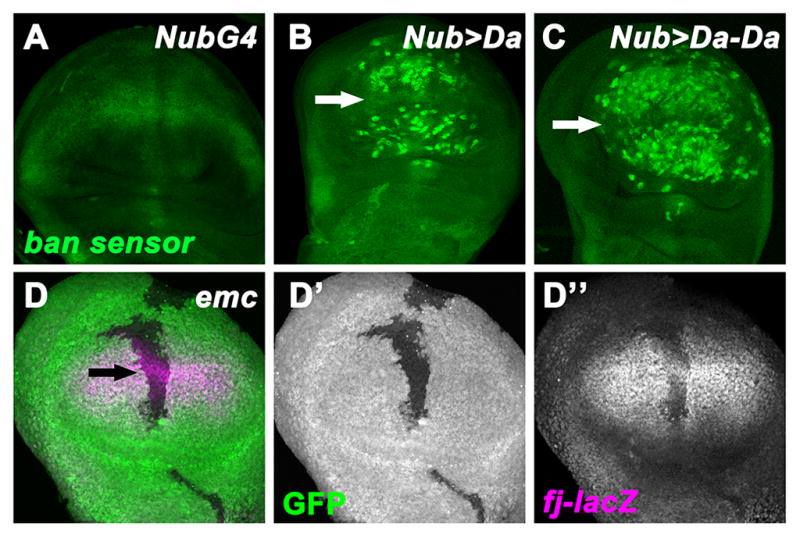
(A–C) Wing imaginal discs of Nub-GAL4 (A), Nub>Da (B), and Nub>Da-Da (C) staining for ban-GFP sensor (green). Activity of ban is negatively reported by the ban-GFP sensor. Note the elevation of ban-GFP sensor in Da-overexpressing cells (wing pouch), except cells near the dorsal-ventral boundary (proneural region, indicated by white arrow). (D–D″) An early-mid third instar wing disc containing emc mutant cells (GFP negative) is visualized for fj-lacZ reporter expression (D″, magenta). Note the decrease levels of fj-lacZ in emc mutant clones. Some emc mutant cells close to the proneural region retain a residual fj-lacZ expression (black arrow).
The physiological role of SWH regulation by Da
During normal development, Da levels are uniform in most uncommitted imaginal disc cells (Cronmiller and Cline, 1987; Bhattacharya and Baker, 2011). If these normal levels contribute to ex transcription and SWH activity, then da null mutant cells would have enhanced growth, like clones mutant for SWH components. When the size of clones homozygous for da null alleles was compared with the reciprocal ‘twin-spot’ clones induced by the same mitotic recombination, however, no difference was seen (Figures 6A–B). Consistent with this finding, EnhIntron3-GFP reporter activity was unaffected in clones of da null mutant cells (Figures 6C and S6A). Therefore da did not appear to act as a brake on the growth rate of normal imaginal disc cells, although we cannot exclude that da may regulate growth through SWH in some other tissue.
Figure 6. Ex removes ectopic High-Da neurons.
(A) Third instar wing imaginal discs containing da10 mutant cells (GFP negative) and their reciprocal twin spot clones (2X GFP staining). (B) Quantification of clone area relative to twin spot area of 10 discs. (C–C′) Third instar wing imaginal discs containing da3 mutant cells (RFP (megenta) negative) visualized for Enhintron3-GFP reporter expression (green). (D–E′) Third instar wing imaginal discs of exe1 (D–D′) and exe1, tub>Da (E–E′) positively marked MARCM clones stained for GFP (green, to mark the clones) and stained for the Senseless (Sens) protein in megenta. Note that ectopic Sens staining in exe1, tub>Da MARCM clones (blue outline in panel E′). (F) Ectopic bristles (white arrow) in adult thorax containing exe1, tub>Da-Da clones. Note that only the clone expressing Da-Da is homozygous for the ex mutant allele. (G) The allelic combination ex1/exe1 can survive to adulthood with minimal growth defects. More than 40% of this genotype differentiate supernumerary thoracic macrochaetae (blue arrows: post-alar bristles; green arows: scutellar bristle). (H) Model. (I) In the normal situation, unspecified progenitor cells proliferate in the imaginal discs. Both da expression and SWH activity maintain basal levels through independent regulatory mechanisms. (II) When emc−/− cells or Da over-expressing cells arise outside the cell-cycle arrested neural regions, elevated levels of Da take over regulation of ex transcription, leading to the activation of the SWH pathway to antagonize Yki activity. Growth and proliferation are blocked, while apoptosis is promoted, preventing such cells contributing to abnormal patterns of neurogenesis. See also Figure S6.
Clones of cells lacking emc are lost from imaginal discs during growth, because of their high Da levels and SWH activity (Garcia Alonso and Garcia-Bellido, 1988; Bhattacharya & Baker, 2011;_Figures 3, 5 in this study). Therefore, the normal level of Emc protein is required for imaginal disc growth, and the signal of inadequate emc activity is high Da. To assess the potential significance of the pathway, we removed ex from Da over-expressing cells to prevent SWH hyperactivity. In exe1; tub>da clones, numerous ectopic sensory organ precursors (SOPs) were detected that were not present in exe1 clones (Figures 6D–E). Previous studies show that da is an especially potent driver of ectopic neurogenesis (Jafar-Nejad et al., 2003). Consistent with this, ectopic bristle sensory organs differentiated in pharate adults (Figures 6F and S6B). We also found hypomorphic ex mutant genotypes that survive with minimal growth defects (Figure 6G). Such genotypes often differentiate supernumerary sensory bristles even without mutation of emc or targeted expression of da (Figure 6G). These data indicate that ex plays a role in fine-tuning neural patterning, and that SWH activity prevents excess neuronal differentiation of cells with inappropriate HLH gene expression by eliminating these cells (Figure 6H).
Discussion
We describe a process that prevents certain mis-specified cells from differentiating into malformed organs. This process creates a requirement for the emc gene in imaginal disc cell growth, since emc loss results in high Da levels that trigger the pathway through transcriptional activation of the ex gene, an upstream regulator of the SWH tumor suppressor pathway. If ex or the downstream SWH genes are mutated, then cells with high Da levels not only survive and grow but produce numerous ectopic neuronal structures. This surveillance function for SWH signalling does not require cell-cell signalling and is distinct from potential roles for SWH in limiting organ growth or preventing tumorigenesis. It may represent an adaptive function for SWH pathway hyperactivity.
Selecting against Progenitor Cells with Incorrect Fate Specification
The heterodimer partners of Da and Emc include proneural bHLH protein that define proneural regions and neural progenitor cells and which are highly regulated in space and time (Modolell and Campuzano, 1998; Quan and Hassan, 2005). Da, by contrast, is expressed ubiquitously and controlled by emc (Bhattacharya and Baker, 2011). Inadequate emc expression permits higher levels of da expression and of Da/bHLH heterodimers, leading to ectopic neural differentiation (Van Doren et al., 1992; Cubas and Modolell, 1992; Bhattacharya and Baker, 2011). Mammalian Id genes are similar feedback regulators of mammalian E proteins (Bhattacharya and Baker, 2011; Schmitz et al., 2012). We have shown here that even if emc expression or its regulation is defective, abnormal neurogenesis is still restrained by SWH signaling that restricts the proliferation and survival of cells with abnormal Da expression. High Da levels directly activate transcription of the ex gene, thereby activating the SWH pathway of tumor suppressors in a cell-autonomous fashion (Figure 6H). Because ex is a feedback inhibitor of SWH signaling that is transcriptionally activated by Yki (Hamaratoglu et al., 2006), ex activation by high Da has the added effect of bypassing feedback control of SWH signaling (Figure 6H), which likely contributes to the efficiency of removal of cells with high Da. Indeed, when ex is removed, cells with high Da are not removed but produce dramatic neural hyperplasia, in which ectopic bristles almost cover a clone in the thoracic epidermis (Figure 6E–F). All these neurogenic defects would be maladaptive in nature, where the pattern of sensory bristles is highly selected (Simpson and Marcellini, 2006).
Our findings suggest that the Da/Emc balance is permissive for normal growth and we have not found evidence for regulation that determines normal organ size or growth rate (Figure 6A, B). By contrast, Da/Emc imbalance outside the normal range in mutant cells triggers the SWH pathway to block growth and remove cells that will otherwise perturb developmental patterning. SWH activation in abnormal development might be analogous to the p53 tumor suppressor, which is inactive in most normal cells, but activated by DNA damage and other stresses (Brady and Attardi, 2010). Interestingly a recent study reported that emc hypomorphic cells, which are less severely affected that emc null cells and can survive in imaginal discs, nevertheless exhibit a growth deficit caused by repression of the cell cycle gene string/cdc25, and that string/cdc25 is repressed directly by abnormally high Da (Andrade-Zapata and Baonza, 2014). Thus there may be multiple, Da-dependent pathways that converge to select against progenitor cells with incorrect cell fate specification.
Implications for development and cancer
Mammalian E proteins and Id proteins are well-established tumor suppressors and proto-oncogenes (Yan et al., 1997; Norton, 2000; Hasskarl and Munger, 2002; Sikder et al., 2003; Perk et al., 2005; Murre, 2005; Iavarone and Lasorella, 2006; Slattery et al., 2008; Kee, 2009). In normal development, E-proteins and Id proteins regulate the coordination of differentiation with cell cycle arrest (Jen et al., 1992; Peverali et al., 1994), and the expansion of mammary epithelial cells in response to pregnancy and lactation (Mori et al., 2000; Parrinello et al., 2001; Itahana et al., 2008; Dong et al., 2011). At least in part, these growth controls relate to the transcriptional activation of cyclin-dependent kinase inhibitor genes by E proteins, such that E proteins are required for cellular senescence, counteracted by Id proteins (Van der Put et al., 2004; Zheng et al., 2004). The senescence mechanisms may not be conserved between mammalian and Drosophila cells (Simcox et al., 2008), but other pathways of tumor suppression by mammalian E proteins exist (Niola et al., 2013), and in certain contexts E proteins can be tumor promoting, and Id proteins tumor suppressive (Schmitz et al., 2012).
The distinctive phenotype of SWH pathway mutations is dramatically enhanced growth and organ size (Pan, 2007; Saucedo and Edgar, 2007). The normal biological functions of the pathway are still debated (Lawrence and Casal, 2013). Reduced SWH activity is implicated in wound healing and regenerative growth (Cai et al., 2010; Karpowicz et al., 2010; Shaw et al., 2010; Zhao et al., 2011; Grusche et al., 2011; Barry et al., 2013). Mice mutant for Mst1, Mst2, Lats1 or Lats2 are tumour prone, suggesting that tumor growth could mimic wound healing or regeneration. Epigenetic silencing of these genes has been reported in human cancer (Takahashi et al., 2005; Jiang et al., 2006; Seidel et al., 2007), where other SWH components are mutated, such as NF2 in neurofibromatosis (Zender et al., 2006; Evans, 2009). Yap is amplified in cancers of liver, colon, lung, ovary (Overholtzer et al., 2006; Steinhardt et al., 2008).
Clearly, SWH activity is normally maintained between a low threshold necessary to prevent hyperplasia, and a high threshold that blocks growth and kills cells. Reduced SWH activity is associated with regenerative responses. In principle, increased SWH might be hyper-activated to eliminate potential tumors, perhaps because of imbalanced expression of E proteins and Id proteins; tumor cells might evolve to evade such a checkpoint. Microarray data from E2A deficiency mice that exhibit high incidence of T-cell leukemia suggest that FRMD6, a mammalian homolog of ex, is an E2A target, which would be consistent with this hypothesis (supplementary data in (Welinder et al., 2011)).
Our work shows directly that in Drosophila hyperactivation of the SWH tumor suppressor pathway can select against cells that express certain developmental errors, which may be adaptive for development. It will be interesting to discover whether SWH signaling can be hyperactivated to remove other kinds of dysfunctional cells besides those expressing inappropriate bHLH protein levels, whether in development or in cancer.
Experimental procedures
Clonal Induction and Clone Size Measurement
Fly culture and cross were performed according to standard procedures at 25 °C except where noted. Immunochemistry and scanning electron microscopy were performed as described (Firth et al., 2006; Baker et al., 2014). Flp-on expression clones were generated by crossing UAS-lines to hs-FLP122; Act5C>CD2> GAL4 UAS-GFP. Mutant clones were made by FLP/FRT-mediated mitotic recombination. FLP was induced by heat shock at 36–48 hr AEL and animals were dissected at late third instar. MARCM was conducted to express UAS-transgenes in mitotic clones of wtsX1, crb82-04 or exe1 mutant cells using ActGAL4 or TubGAL4. Heat shocks were performed at 24–48 hr AEL and incubated at 26.5°C until animals were dissected. Nub>da experiments were performed at 18°C to minimize lethality. Clone sizes and perimeters were measured in microns using NIH Image J.
Quantitative RT-PCR
Eyg-GAL4>UAS-GFP (control) and eyg-GAL4> UAS-Da salivary gland were dissected in PBS. Total RNA was isolated using the Zymo Research Micro RNA isolation kit and DNA-Free RNA kit (Zymo Research). First Strand cDNA Synthesis Kit for RT-PCR (AMV; Roche) and random hexamer oligo-p(dT)15 primers were used to produce cDNAs from the extracted total RNA (1 μg). To measure mRNA levels, Real-time qPCR was performed with SYBR Green Master PCR Mix (ABI) using the ABI 7900HT Detection System. All reactions were performed three times. The relative amount of specific mRNAs under each condition was calculated after normalization to the rp49 transcript. rp49-5′: TACAGGCCCAAGATCGTGAA, rp49-3′: ACGTTGTGCACCAGGAACTT; ex-5′: CAGCAGCAGCCGAAAACCT, ex-3′: TTGGGCCATATTTTGAGAGC; mer-5′: AAGCACGACCTGGAGAAGAA, mer-3′:AGGCTATCCGTGGAGGACTT; hpo-5′:GGAGTCGAACTTGGGCACTA, hpo-3′: GCTGCTGCTGTTGTTGTTGT. The statistical significance of the difference was assessed using a Student’s t-test with significance at P<0.05.
Chromatin Immunoprecipitation
Wing imaginal discs were dissected from late third instar larvae in cold PBS and fixed for 20 min at room temperature in 1.8% formaldehyde. After quenching in 0.125M Glycine, the discs were washed twice in cold PBS containing 0.01% Triton and complete protease inhibitor cocktail (Roche). Incubation of the discs with cold cell lysis buffer (5 mM PIPES, 85 mM KCl, 0.5% NP-40) for 10 min on ice and centrifugation 5 min at 4°C. Pellet was resuspended with nuclei lysis buffer (50 mM Tris-Cl, 10 mM EDTA, 1% SDS and complete protease inhibitor cocktail) to perform sonication. Soluble chromatin was transfer to a new tube after centrifugation, and 10% was removed for input. Chromatin was precleared with protein A/G agarose beads (Santa Cruz) for 1 hr, then incubated overnight with HA probe (F-7) antibody (Santa Cruz) or control IgG (normal mouse IgG, Santa Cruz) in ChIP dilution buffer (16.7 mM Tris-Cl, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100, complete protease inhibitor cocktail). Antibody-chromatin complexes were pulled down with beads (Invitrogen) for 3 hr at 4°C. Beads were washed five times in high salt wash buffer (50 mM HEPES, 500 mM NaCl, 1 mM EDTA, 0.1%SDS, 1% Triton X-100, 0.1% deoxycholate), and twice in TE buffer. Chromatin was eluted in ChIP elution buffer (1% SDS, 0.1M NaHCO3) supplemented with 1 uL of proteinase K (20 ug/uL) and RNase. To reverse crosslinks, eluted materials were incubated at 65°C over 6 hr. PCR purification kit (Qiagen) was used to clean up DNA. Real-time PCR analysis was performed on ABI 7900HT instrument using SYBR Green Master PCR Mix (ABI). Results were quantified using the delta Ct method, normalizing to the input samples.
Supplementary Material
Figure S1. Related to Figure 1
(A–B) UAS-GFP expressed under the control of eyg-GAL4 in third instar eye imaginal discs (A) and salivary glands (B). (C) An adult eye of wild-type. (D) An adult eye of eyg>Da flies. Note the reduced size. (E) An adult eye of eyg>emc dsRNA flies. Note the reduction of eye size and ectopic bristles. The phenotype is more variable than for eyg>da. (F) eygCD>Da, heterozygous for emcAP6. The small eye is further reduced and sometimes absent. (G) Adult eye of GMR>Da (H) Adult eye of GMR>Da, wtsX1/+. (I) eygCD>Da, heterozygous for H99 deficiency. (J) Overexpression of diap1 in eygCD>Da. (K) Overexpression of dominant negative dronc in eygCD>Da. (L) eygCD>Da, heterozygous for darkK11502dark82. (M) eygCD>Da, heterozygous for E2f107172. This ‘headless’ phenotype is the most extreme observed. (N) eygCD>Da, heterozygous for CycEAR95. (O) eygCD>Da, heterozygous for stg4. (P) eygCD>Da, heterozygous for wee1ES1.
Figure S2. Related to Figure 2
(A–H) Third instar wing imaginal discs with GFP-expressing MARCM clones. (A) control (B) wtsX1homozygous (C) wtsX1 homozygous, act>Da (D) act>Da. Clones of wtsX1 or wtsX1, act>Da grow well, whereas act>Da clones were rarely recovered. (E) control (F), ftNY1 homozygous (G), ftNY1 homozygous, tub>Da (H) tub>Da. Clones of ftNY1 or ftNY1, tub>Da grew well throughout the discs, whereas tub>Da clones were rarely recovered. (I) Quantification of results. 10 discs of each genotype were analyzed. Area of clones was normalized to the total wing disc area. Data are represented as mean±SEM. NS, not significant. (J) Eye disc with wts homozygous clones stained for GFP (green) and Da (magenta). (J′, J″) separate GFP and Da channels.
Figure S3. Related to Figure 3
(A–B) Wing imaginal discs (A) or salivary glands (B) containing control clones (ActGal4>GFP+P35, GFP positive, green) and stained for ex-LacZ reporter expression (magenta). Unlike Da-overexpressing clones (Figure 3), no elevation is observed in ActGal4>GFP+P35 clones. (C–D) Wing imaginal discs of NubGAL4 (C) and Nub>Da (D), staining for Ex antibody (red). (E) Salivary glands containing Da-overexpressing clones staining for Ex antibody (same specimen as Figure 3D). Note the elevation of Ex protein levels in Da-overexpressing cells (D and E). (F–G) Wing imaginal discs of NubGAL4 (F) and Nub>Da (G), staining for Dcp-1 antibody to identify dying cells (red). (H–K) Adult wing size after Da and p35 over-expression. (H) Control NubGAL4 wing. (I) Nub>Da reduces wing size. (J) Expression of p35 partially restores Nub>Da wing size. (K) Nub>p35 wing. (L) Quantification of the wing size after Da and p35 over-expression.
Figure S4. Related to Figure 4
(A–C) Adult eyes of eyg>Da-Da flies. The size reduction is greater than for Da and can be extreme (B) or remove the head (C). (D–E) Wing discs of Nub>Ac (D) and Nub>Sc (E), staining for Enh intron3-GFP reporter expression. Enh intron3-GFP is upregulated. (F) ChIP analysis of the EnhIntron3 enhancer. Anti-histone H3 (HA IP) or IgG antibodies wer used to precipitate chromatin from wing discs of NubGAL4 and Nub>UAS-HA-Da. Quantitative PCR was done on regions A–G (see panel 4A). Graph shows the relative fold changes of anti-histone H3 compared to IgG. Results represent mean±SEM (n=3).
Figure S5. Related to Figure 5
(A) Third instar wing disc containing wild-type clones in a Minute background (GFP negative, A′). Note similar fj-lacZ expression levels between wild-type cells (GFP negative) and Minute cells (GFP positive)(magenta; A″). (B) Wing imaginal disc containing Da-overexpressing clones (ActGal4>Da, RFP positive, red). Note ban-GFP sensor is elevated in Da-overexpressing clones (green, B″). (C) Wing imaginal disc containing Da-overexpressing clones (ActGal4>Da, GFP positive, green). Note the decrease levels of fj-lacZ in Da-overexpressing clones (red, C″). (D) Wing imaginal disc containing YkiS168A-overexpressing clones (ActGal4>YkiS168A, GFP positive, green). Note that fj-lacZ is elevated by active Yki (red, D″). (E) Wing imaginal disc containing clones coexpressing Da and YkiS168A (ActGal4>Da+YkiS168A, GFP positive, green). Note fj-lacZ (red, E″) and large clones. All crosses in B–E maintained at 18. (F) Wing imaginal disc containing emc mutant cells (GFP negative, F′). diap1-lacZ reporter expression (F″, magenta) reduction in emc mutant clones. (G) Third instar wing disc containing wild-type control clones (GFP negative). Note that there is no difference of diap1-lacZ expression (magenta, G′).
Figure S6. Related to Figure 6
(A) Third instar eye imaginal discs containing da3 mutant cells (RFP negative) visualized for Enhintron3-GFP reporter expression (green). (B) Ectopic bristles (white arrows) in adult thorax containing exe1, tub>Da clones.
Table S1. Summary of deficiency screen for dominant modifiers. Related to Figure 1.
414 deficiency stocks from the DrosDel and Exelixis collections were screened to identify genomic regions that exhibited a modifying effect on eyg>Da phenotype. From this screen, a total of 38 genomic regions were identified based on their modifier capability. 30 genomic regions showed suppression and 8 genomic regions showed enhancement on eyg>Da phenotype.
Table S2. A subset of cell cycle/cell death genes genetically interact with eyg>da. Related to Figure 1.
Key: +++, strong enhancement; ++ moderate enhancement; + mild enhancement; −−− strong suppression; −− moderate suppression; − mild suppression.
Acknowledgments
We thank Claude Desplan, Georg Halder, Iswar Hariharan, Laura Johnston Duojia Pan, and Gary Struhl for fly strains, and Andreas Jenny, Julie Secombe and Andrew Tomlinson for comments on the manuscript. Confocal and Electron microscopy performed in the Analytical imaging Facility at AECOM supported by the NCI (P30CA013330). Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Supported by NIH grant GM047892, and by an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology and Visual Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrade-Zapata I, Baonza A. The bHLH factors extramacrochaetae and daughterless control cell cycle in Drosophila imaginal discs through the transcriptional regulation of the Cdc25 phosphatase string. PLoS Genet. 2014;10:e1004233. doi: 10.1371/journal.pgen.1004233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE, Li K, Quiquand M, Ruggiero R, Wang LH. Eye development. Methods. 2014;68:252–259. doi: 10.1016/j.ymeth.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726, 728, 730, 732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Barry ER, Morikawa T, Butler BL, Shrestha K, de la Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, Kuo CJ, Camargo FD. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Baker NE. A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell. 2011;147:881–892. doi: 10.1016/j.cell.2011.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggiano JC, Fehon RG. Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev Cell. 2012;22:695–702. doi: 10.1016/j.devcel.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt W, Chen CL, Chen Q, Sudol M, McNeill H, Pan D, Kopp A, Halder G. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene. 2014;33:1218–1228. doi: 10.1038/onc.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CA, Attardi LD. p53 at a glance. J Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, Pan D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano S. Emc, a negative HLH regulator with multiple functions in Drosophila development. Oncogene. 2001;20:8299–8307. doi: 10.1038/sj.onc.1205162. [DOI] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- Cook RK, Christensen SJ, Deal JA, Coburn RA, Deal ME, Gresens JM, Kaufman TC, Cook KR. The generation of chromosomal deletions to provide extensive coverage and subdivision of the Drosophila melanogaster genome. Genome Biol. 2012;13:R21. doi: 10.1186/gb-2012-13-3-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronmiller C, Cline TW. The Drosophila sex determination gene daughterless has different functions in the germ line versus the soma. Cell. 1987;48:479–487. doi: 10.1016/0092-8674(87)90198-x. [DOI] [PubMed] [Google Scholar]
- Cubas P, Modolell J. The extramacrochaetae gene provides information for sensory organ patterning. EMBO J. 1992;11:3385–3393. doi: 10.1002/j.1460-2075.1992.tb05417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Baonza A, Garcia-Bellido A. Behavior of extramacrochaetae mutant cells in the morphogenesis of the Drosophila wing. Mech Dev. 1995;53:209–221. doi: 10.1016/0925-4773(95)00436-5. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Huang S, Caikovski M, Ji S, McGrath A, Custorio MG, Creighton CJ, Maliakkal P, Bogoslovskaia E, Du Z, Zhang X, Lewis MT, Sablitzky F, Brisken C, Li Y. ID4 regulates mammary gland development by suppressing p38MAPK activity. Development. 2011;138:5247–5256. doi: 10.1242/dev.069203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG. Neurofibromatosis 2 [Bilateral acoustic neurofibromatosis, central neurofibromatosis, NF2, neurofibromatosis type II] Genet Med. 2009;11:599–610. doi: 10.1097/GIM.0b013e3181ac9a27. [DOI] [PubMed] [Google Scholar]
- Feng Y, Irvine KD. Fat and expanded act in parallel to regulate growth through warts. Proc Natl Acad Sci U S A. 2007;104:20362–20367. doi: 10.1073/pnas.0706722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth LC, Li W, Zhang H, Baker NE. Analyses of RAS regulation of eye development in Drosophila melanogaster. Methods Enzymol. 2006;407:711–721. doi: 10.1016/S0076-6879(05)07056-4. [DOI] [PubMed] [Google Scholar]
- Garcia Alonso LA, Garcia-Bellido A. Extramacrochaetae, a transacting gene of the achaete-scute complex of Drosophila involved in cell communication. Roux Arch Dev Biol. 1988;197:328–338. doi: 10.1007/BF00375952. [DOI] [PubMed] [Google Scholar]
- Genevet A, Tapon N. The Hippo pathway and apico-basal cell polarity. Biochem J. 2011;436:213–224. doi: 10.1042/BJ20110217. [DOI] [PubMed] [Google Scholar]
- Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding SE, zur LP, Jarman AP. amos, a proneural gene for Drosophila olfactory sense organs that is regulated by lozenge. Neuron. 2000;25:69–78. doi: 10.1016/s0896-6273(00)80872-7. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Grusche FA, Degoutin JL, Richardson HE, Harvey KF. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Dev Biol. 2011;350:255–266. doi: 10.1016/j.ydbio.2010.11.020. [DOI] [PubMed] [Google Scholar]
- Grusche FA, Richardson HE, Harvey KF. Upstream regulation of the hippo size control pathway. Curr Biol. 2010;20:R574–R582. doi: 10.1016/j.cub.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hasskarl J, Munger K. Id proteins--tumor markers or oncogenes? Cancer Biol Ther. 2002;1:91–96. doi: 10.4161/cbt.50. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Huang ML, Hsu CH, Chien CT. The proneural gene amos promotes multiple dendritic neuron formation in the Drosophila peripheral nervous system. Neuron. 2000;25:57–67. doi: 10.1016/s0896-6273(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Iavarone A, Garg P, Lasorella A, Hsu J, Israel MA. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- Iavarone A, Lasorella A. ID proteins as targets in cancer and tools in neurobiology. Trends Mol Med. 2006;12:588–594. doi: 10.1016/j.molmed.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Itahana Y, Piens M, Fong S, Singh J, Sumida T, Desprez PY. Expression of Id and ITF-2 genes in the mammary gland during pregnancy. Biochem Biophys Res Commun. 2008;372:826–830. doi: 10.1016/j.bbrc.2008.05.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafar-Nejad H, Acar M, Nolo R, Lacin H, Pan H, Parkhurst SM, Bellen HJ. Senseless acts as a binary switch during sensory organ precursor selection. Genes Dev. 2003;17:2966–2978. doi: 10.1101/gad.1122403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Jen Y, Weintraub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev. 1992;6:1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Li X, Hu J, Zhou W, Jiang Y, Li G, Lu D. Promoter hypermethylation-mediated down-regulation of LATS1 and LATS2 in human astrocytoma. Neurosci Res. 2006;56:450–458. doi: 10.1016/j.neures.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Jukam D, Desplan C. Binary regulation of Hippo pathway by Merlin/NF2, Kibra, Lgl, and Melted specifies and maintains postmitotic neuronal fate. Dev Cell. 2011;21:874–887. doi: 10.1016/j.devcel.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003;17:1090–1100. doi: 10.1101/gad.1054603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A, Iavarone A, Israel MA. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar AB, Davis RL, Wright WE, Kadesch T, Murre C, Voronova A, Baltimore D, Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991;66:305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Casal J. The mechanisms of planar cell polarity, growth and the Hippo pathway: some known unknowns. Dev Biol. 2013;377:1–8. doi: 10.1016/j.ydbio.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, Knowles DW, Stapleton M, Bickel P, Biggin MD, Eisen MB. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 2009;10:R80. doi: 10.1186/gb-2009-10-7-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond J, Strelets VB, Wilson RJ. FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41:D751–D757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 2011;21:727–735. doi: 10.1016/j.tcb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Modolell J, Campuzano S. The achaete-scute complex as an integrating device. Int J Dev Biol. 1998;42:275–282. [PubMed] [Google Scholar]
- Mori S, Nishikawa SI, Yokota Y. Lactation defect in mice lacking the helix-loop-helix inhibitor Id2. EMBO J. 2000;19:5772–5781. doi: 10.1093/emboj/19.21.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Niola F, Zhao X, Singh D, Sullivan R, Castano A, Verrico A, Zoppoli P, Friedmann-Morvinski D, Sulman E, Barrett L, Zhuang Y, Verma I, Benezra R, Aldape K, Iavarone A, Lasorella A. Mesenchymal high-grade glioma is maintained by the ID-RAP1 axis. J Clin Invest. 2013;123:405–417. doi: 10.1172/JCI63811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, Makita R, Kurihara H, Morin-Kensicki EM, Nojima H, Rossant J, Nakao K, Niwa H, Sasaki H. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(Pt 22):3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Parrinello S, Lin CQ, Murata K, Itahana Y, Singh J, Krtolica A, Campisi J, Desprez PY. Id-1, ITF-2, and Id-2 comprise a network of helix-loop-helix proteins that regulate mammary epithelial cell proliferation, differentiation, and apoptosis. J Biol Chem. 2001;276:39213–39219. doi: 10.1074/jbc.M104473200. [DOI] [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- Peverali FA, Ramqvist T, Saffrich R, Pepperkok R, Barone MV, Philipson L. Regulation of G1 progression by E2A and Id helix-loop-helix proteins. EMBO J. 1994;13:4291–4301. doi: 10.1002/j.1460-2075.1994.tb06749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan XJ, Hassan BA. From skin to nerve: flies, vertebrates and the first helix. Cell Mol Life Sci. 2005;62:2036–2049. doi: 10.1007/s00018-005-5124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Camargo FD. The Hippo signaling pathway and stem cell biology. Trends Cell Biol. 2012;22:339–346. doi: 10.1016/j.tcb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roote J, Russell S. Toward a complete Drosophila deficiency kit. Genome Biol. 2012;13:149. doi: 10.1186/gb-2012-13-3-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, Morley T, Chan YS, Blows F, Coulson D, Reuter G, Baisch H, Apelt C, Kauk A, Rudolph T, Kube M, Klimm M, Nickel C, Szidonya J, Maroy P, Pal M, Rasmuson-Lestander A, Ekstrom K, Stocker H, Hugentobler C, Hafen E, Gubb D, Pflugfelder G, Dorner C, Mechler B, Schenkel H, Marhold J, Serras F, Corominas M, Punset A, Roote J, Russell S. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177:615–629. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E, Blows F, Ashburner M, Bautista-Llacer R, Coulson D, Drummond J, Webster J, Gubb D, Gunton N, Johnson G, O’Kane CJ, Huen D, Sharma P, Asztalos Z, Baisch H, Schulze J, Kube M, Kittlaus K, Reuter G, Maroy P, Szidonya J, Rasmuson-Lestander A, Ekstrom K, Dickson B, Hugentobler C, Stocker H, Hafen E, Lepesant JA, Pflugfelder G, Heisenberg M, Mechler B, Serras F, Corominas M, Schneuwly S, Preat T, Roote J, Russell S. The DrosDel collection: a set of P-element insertions for generating custom chromosomal aberrations in Drosophila melanogaster. Genetics. 2004;167:797–813. doi: 10.1534/genetics.104.026658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- Schmitz R, Young RM, Ceribelli M, Jhavar S, Xiao W, Zhang M, Wright G, Shaffer AL, Hodson DJ, Buras E, Liu X, Powell J, Yang Y, Xu W, Zhao H, Kohlhammer H, Rosenwald A, Kluin P, Muller-Hermelink HK, Ott G, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Ogwang MD, Reynolds SJ, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pittaluga S, Wilson W, Waldmann TA, Rowe M, Mbulaiteye SM, Rickinson AB, Staudt LM. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MC, Halder G. Regulation of the Hippo pathway by cell architecture and mechanical signals. Semin Cell Dev Biol. 2012;23:803–811. doi: 10.1016/j.semcdb.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Schwank G, Tauriello G, Yagi R, Kranz E, Koumoutsakos P, Basler K. Antagonistic growth regulation by Dpp and Fat drives uniform cell proliferation. Dev Cell. 2011;20:123–130. doi: 10.1016/j.devcel.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Seidel C, Schagdarsurengin U, Blumke K, Wurl P, Pfeifer GP, Hauptmann S, Taubert H, Dammann R. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46:865–871. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- Shaw RL, Kohlmaier A, Polesello C, Veelken C, Edgar BA, Tapon N. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Simcox A, Mitra S, Truesdell S, Paul L, Chen T, Butchar JP, Justiniano S. Efficient genetic method for establishing Drosophila cell lines unlocks the potential to create lines of specific genotypes. PLoS Genet. 2008;4:e1000142. doi: 10.1371/journal.pgen.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P, Marcellini S. The origin and evolution of stereotyped patterns of macrochaetes on the nota of cyclorraphous Diptera. Heredity (Edinb ) 2006;97:148–156. doi: 10.1038/sj.hdy.6800874. [DOI] [PubMed] [Google Scholar]
- Slattery C, Ryan MP, McMorrow T. E2A proteins: regulators of cell phenotype in normal physiology and disease. Int J Biochem Cell Biol. 2008;40:1431–1436. doi: 10.1016/j.biocel.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Staley BK, Irvine KD. Hippo signaling in Drosophila: recent advances and insights. Dev Dyn. 2012;241:3–15. doi: 10.1002/dvdy.22723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, Noguchi S. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11:1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler DM, Li W, Zhuo N, Pellock B, Baker NE. Genes affecting cell competition in Drosophila. Genetics. 2007;175:643–657. doi: 10.1534/genetics.106.061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Van der Put E, Frasca D, King AM, Blomberg BB, Riley RL. Decreased E47 in senescent B cell precursors is stage specific and regulated posttranslationally by protein turnover. J Immunol. 2004;173:818–827. doi: 10.4049/jimmunol.173.2.818. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Ellis HM, Posakony JW. The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- Van Doren M, Powell PA, Pasternak D, Singson A, Posakony JW. Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- Welinder E, Mansson R, Mercer EM, Bryder D, Sigvardsson M, Murre C. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci U S A. 2011;108:17402–17407. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci U S A. 2008;105:14897–14902. doi: 10.1073/pnas.0805201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Yan W, Young AZ, Soares VC, Kelley R, Benezra R, Zhuang Y. High incidence of T-cell tumors in E2A-null mice and E2A/Id1 double-knockout mice. Mol Cell Biol. 1997;17:7317–7327. doi: 10.1128/mcb.17.12.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecca M, Struhl G. A feed-forward circuit linking wingless, fat-dachsous signaling, and the warts-hippo pathway to Drosophila wing growth. PLoS Biol. 2010;8:e1000386. doi: 10.1371/journal.pbio.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, Mu D, Lucito R, Powers S, Lowe SW. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Wang H, Xue L, Zhang Z, Tong T. Regulation of cellular senescence and p16(INK4a) expression by Id1 and E47 proteins in human diploid fibroblast. J Biol Chem. 2004;279:31524–31532. doi: 10.1074/jbc.M400365200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figure 1
(A–B) UAS-GFP expressed under the control of eyg-GAL4 in third instar eye imaginal discs (A) and salivary glands (B). (C) An adult eye of wild-type. (D) An adult eye of eyg>Da flies. Note the reduced size. (E) An adult eye of eyg>emc dsRNA flies. Note the reduction of eye size and ectopic bristles. The phenotype is more variable than for eyg>da. (F) eygCD>Da, heterozygous for emcAP6. The small eye is further reduced and sometimes absent. (G) Adult eye of GMR>Da (H) Adult eye of GMR>Da, wtsX1/+. (I) eygCD>Da, heterozygous for H99 deficiency. (J) Overexpression of diap1 in eygCD>Da. (K) Overexpression of dominant negative dronc in eygCD>Da. (L) eygCD>Da, heterozygous for darkK11502dark82. (M) eygCD>Da, heterozygous for E2f107172. This ‘headless’ phenotype is the most extreme observed. (N) eygCD>Da, heterozygous for CycEAR95. (O) eygCD>Da, heterozygous for stg4. (P) eygCD>Da, heterozygous for wee1ES1.
Figure S2. Related to Figure 2
(A–H) Third instar wing imaginal discs with GFP-expressing MARCM clones. (A) control (B) wtsX1homozygous (C) wtsX1 homozygous, act>Da (D) act>Da. Clones of wtsX1 or wtsX1, act>Da grow well, whereas act>Da clones were rarely recovered. (E) control (F), ftNY1 homozygous (G), ftNY1 homozygous, tub>Da (H) tub>Da. Clones of ftNY1 or ftNY1, tub>Da grew well throughout the discs, whereas tub>Da clones were rarely recovered. (I) Quantification of results. 10 discs of each genotype were analyzed. Area of clones was normalized to the total wing disc area. Data are represented as mean±SEM. NS, not significant. (J) Eye disc with wts homozygous clones stained for GFP (green) and Da (magenta). (J′, J″) separate GFP and Da channels.
Figure S3. Related to Figure 3
(A–B) Wing imaginal discs (A) or salivary glands (B) containing control clones (ActGal4>GFP+P35, GFP positive, green) and stained for ex-LacZ reporter expression (magenta). Unlike Da-overexpressing clones (Figure 3), no elevation is observed in ActGal4>GFP+P35 clones. (C–D) Wing imaginal discs of NubGAL4 (C) and Nub>Da (D), staining for Ex antibody (red). (E) Salivary glands containing Da-overexpressing clones staining for Ex antibody (same specimen as Figure 3D). Note the elevation of Ex protein levels in Da-overexpressing cells (D and E). (F–G) Wing imaginal discs of NubGAL4 (F) and Nub>Da (G), staining for Dcp-1 antibody to identify dying cells (red). (H–K) Adult wing size after Da and p35 over-expression. (H) Control NubGAL4 wing. (I) Nub>Da reduces wing size. (J) Expression of p35 partially restores Nub>Da wing size. (K) Nub>p35 wing. (L) Quantification of the wing size after Da and p35 over-expression.
Figure S4. Related to Figure 4
(A–C) Adult eyes of eyg>Da-Da flies. The size reduction is greater than for Da and can be extreme (B) or remove the head (C). (D–E) Wing discs of Nub>Ac (D) and Nub>Sc (E), staining for Enh intron3-GFP reporter expression. Enh intron3-GFP is upregulated. (F) ChIP analysis of the EnhIntron3 enhancer. Anti-histone H3 (HA IP) or IgG antibodies wer used to precipitate chromatin from wing discs of NubGAL4 and Nub>UAS-HA-Da. Quantitative PCR was done on regions A–G (see panel 4A). Graph shows the relative fold changes of anti-histone H3 compared to IgG. Results represent mean±SEM (n=3).
Figure S5. Related to Figure 5
(A) Third instar wing disc containing wild-type clones in a Minute background (GFP negative, A′). Note similar fj-lacZ expression levels between wild-type cells (GFP negative) and Minute cells (GFP positive)(magenta; A″). (B) Wing imaginal disc containing Da-overexpressing clones (ActGal4>Da, RFP positive, red). Note ban-GFP sensor is elevated in Da-overexpressing clones (green, B″). (C) Wing imaginal disc containing Da-overexpressing clones (ActGal4>Da, GFP positive, green). Note the decrease levels of fj-lacZ in Da-overexpressing clones (red, C″). (D) Wing imaginal disc containing YkiS168A-overexpressing clones (ActGal4>YkiS168A, GFP positive, green). Note that fj-lacZ is elevated by active Yki (red, D″). (E) Wing imaginal disc containing clones coexpressing Da and YkiS168A (ActGal4>Da+YkiS168A, GFP positive, green). Note fj-lacZ (red, E″) and large clones. All crosses in B–E maintained at 18. (F) Wing imaginal disc containing emc mutant cells (GFP negative, F′). diap1-lacZ reporter expression (F″, magenta) reduction in emc mutant clones. (G) Third instar wing disc containing wild-type control clones (GFP negative). Note that there is no difference of diap1-lacZ expression (magenta, G′).
Figure S6. Related to Figure 6
(A) Third instar eye imaginal discs containing da3 mutant cells (RFP negative) visualized for Enhintron3-GFP reporter expression (green). (B) Ectopic bristles (white arrows) in adult thorax containing exe1, tub>Da clones.
Table S1. Summary of deficiency screen for dominant modifiers. Related to Figure 1.
414 deficiency stocks from the DrosDel and Exelixis collections were screened to identify genomic regions that exhibited a modifying effect on eyg>Da phenotype. From this screen, a total of 38 genomic regions were identified based on their modifier capability. 30 genomic regions showed suppression and 8 genomic regions showed enhancement on eyg>Da phenotype.
Table S2. A subset of cell cycle/cell death genes genetically interact with eyg>da. Related to Figure 1.
Key: +++, strong enhancement; ++ moderate enhancement; + mild enhancement; −−− strong suppression; −− moderate suppression; − mild suppression.