Figure 4.
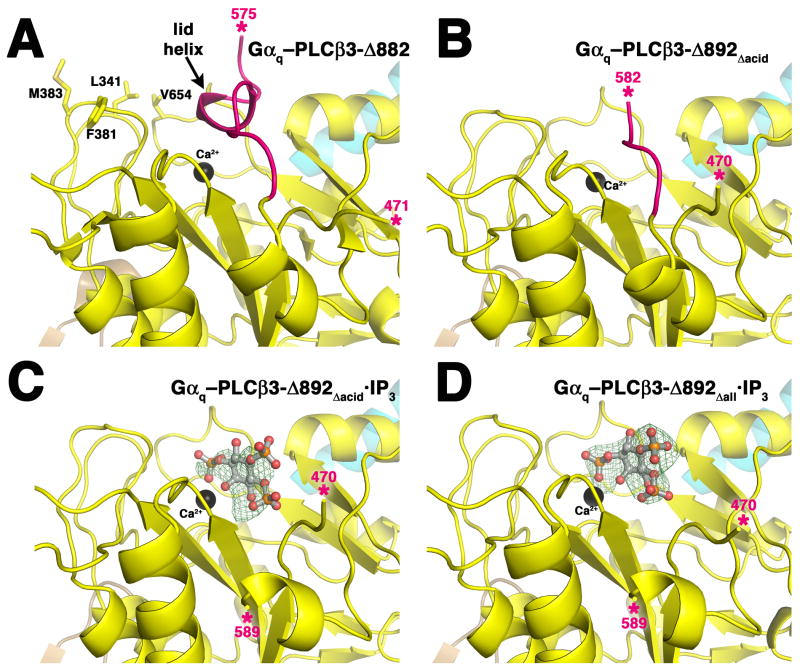
Structural characterization of PLCβ3 variants. PLCβ3 variants are colored as in Figure 1A, and the observed ends of the X–Y linker are marked with pink asterisks. The Hα2′ helix is shown in cyan. (A) Active site of the Gαq–PLCβ3-Δ882 complex (PDB entry 3OHM). Residues Leu341, Phe381, Met383, and Val654 form the hydrophobic ridge, which anchors the active site at the membrane for catalysis (Essen et al., 1997; Lyon and Tesmer, 2013). The lid helix at the C-terminal end of the X–Y linker physically occludes the active site. (B) The active site of the Gαq–PLCβ3-Δ892Δacid complex is open. Residues that would form the lid helix are present in this variant, but deletion of the acidic stretch leads to their disorder. (C) Active site of the Gαq–PLCβ3-Δ892Δacid·IP3 complex. Electron density is observed for most atoms of the ligand, as seen in a 3σ |Fo|-|Fc| omit map (green wire cage). Electron density for the X–Y linker is not observed until residue 589. (D) Active site of the Gαq–PLCβ3-Δ892Δall·IP3 complex. Strong 3σ |Fo|-|Fc| omit map density is observed for the entire IP3 molecule. Deletion of the entire X–Y linker in this variant has no effect on the position of the bound ligand. See also Supplemental Figure 4.