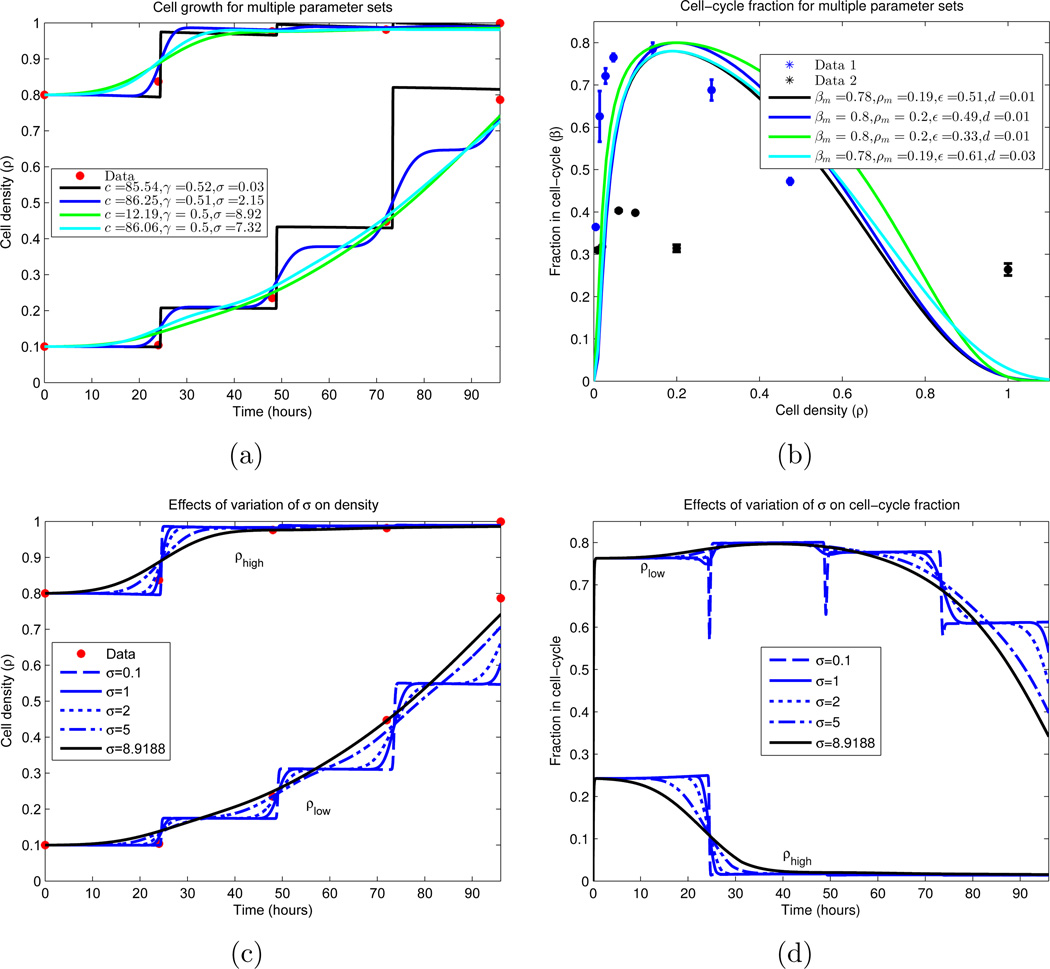
Figure 8.
Cellular growth dynamics for varying parameter sets. (a) A nonlinear least-squares algorithm was used to locate parameter values which minimize the difference between the experimental data and the IDE (equations (18)–(20), together with (9)–(10)). Due to the sparsity of the data (red), multiple local minimums were found. We note that all curves plotted have residuals with ℓ2 norm ∈ [0.0583, 0.0980], and are displayed in descending order by this measure. Similar plot styles in (a) and (b) correspond to the same parameter set. (a) Growth of cellular density in time; (b) Division (β) and death fractions (d = β(1)) for the fitted parameter sets.
(c)–(d) Dependence of IDE solutions on cell-cycle standard deviation σ. σ is varied, while all other parameters are fixed from a local minimizing set located in (a) and (b). We plot the global density growth and cell-cycle fraction for each case. (c) Cellular density ρ for σ variation; (d) Proliferative compartment for σ variation.