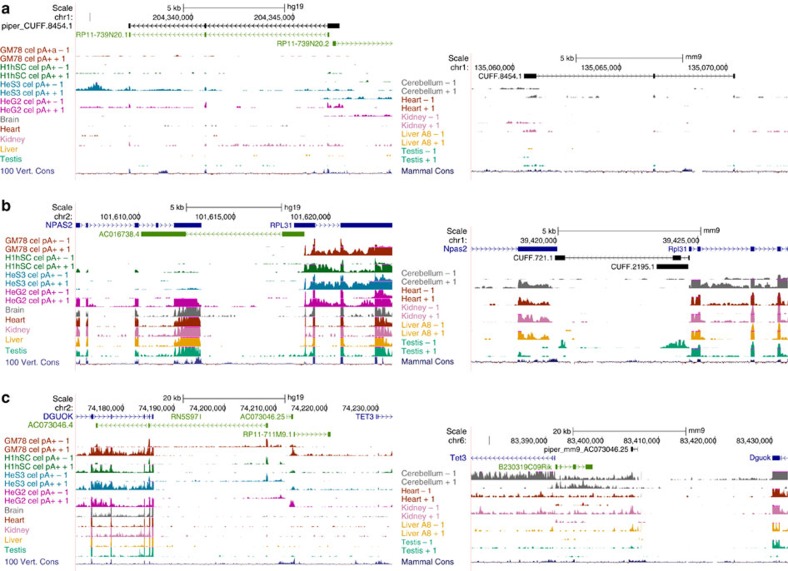
Figure 1. Examples of extensions to the mouse annotation (left: human; right: mouse).
(a) The novel intergenic transcript model CUFF.8454.1 has been inferred from mouse RNA-seq data in a region of the mouse genome without gene annotations. Mapping of this transcript model to the human genome reveals that it is the homologue of the lncRNA RP11-739N20.1 annotated in the human genome. (b) Two mouse transcript models, CUFF.721.1 and CUFF.2195.1, have predicted antisense to two neighbouring protein-coding loci (Npas2 and RPL31). RNA-seq shows that their expression is restricted to testis. CUFF.721.1 is the likely homologue of the annotated human ACO16738.4, which is antisense and overlaps the human orthologue of Npas2 and RPL31. In human, ACO16738.4 appears to be also expressed specifically in testis, although this is not conclusive, as we lack stranded RNA-seq data. (c) The mapping of the human AC0703046.25 lncRNA to the mouse genome identified a potential mouse transcript in the syntenic region between the DGUOK and the TET3 genes (piper_mm9_AC073046.25). This transcript was not included in our predicted models, but it has strong support by RNA-seq data, specifically in cerebellum, kidney and liver. Tissue RNA-seq data in human are from Ilumina Body Map HBM. Plots are UCSC browser screenshots where novel mouse models are indicated in black, human gencode annotation in blue, green and red, and mouse CSHL and human HBM RNA-seq signal in different colours at the bottom. Annotated genes are represented by the longest transcript.